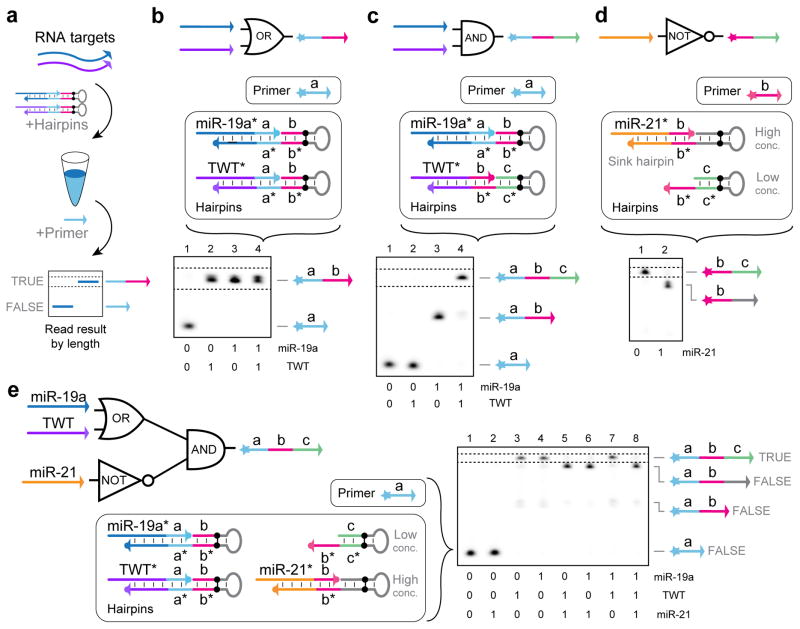
Figure 6. Logic computation with PER.
(a) Operational schematic for evaluating logic expressions with RNA inputs. (b) miR-19a OR TWT gate reaction components and a PAGE denaturing gel depicting transcript production in response to the different RNA inputs. True outputs are read by looking for transcripts of a particular length, indicated by the dotted lines. Reaction setup and PAGE denaturing gel results are also shown for (c) miR-19a AND TWT, (d) NOT miR-21, and (e) (miR-19a OR TWT) AND (NOT miR-21). In all experiments, RNA targets and hairpin protectors were fixed at 250 nM final concentrations. Hairpins for part (b) were at 100 nM. Part (c) used 100 nM and 200 nM for the miR-19a and TWT hairpins, respectively. Part (d) held the miR-21 sink hairpin at a concentration of 150 nM, and the slow unprotected hairpin was held at 30 nM. Part (e) used 100 nM miR-19a and TWT hairpins, 200 nM miR-21 sink hairpin, and 40 nM slow unprotected hairpin. After 15 minutes of pre-incubation at 37 °C with the targets and protected hairpins, primers were introduced and incubated for 2 hours, 2 hours, 3 hours, and 5 hours for parts (b) through (e), respectively. All reactions used 10 μM each of dATP, dTTP, and dCTP. See Methods section for additional details.