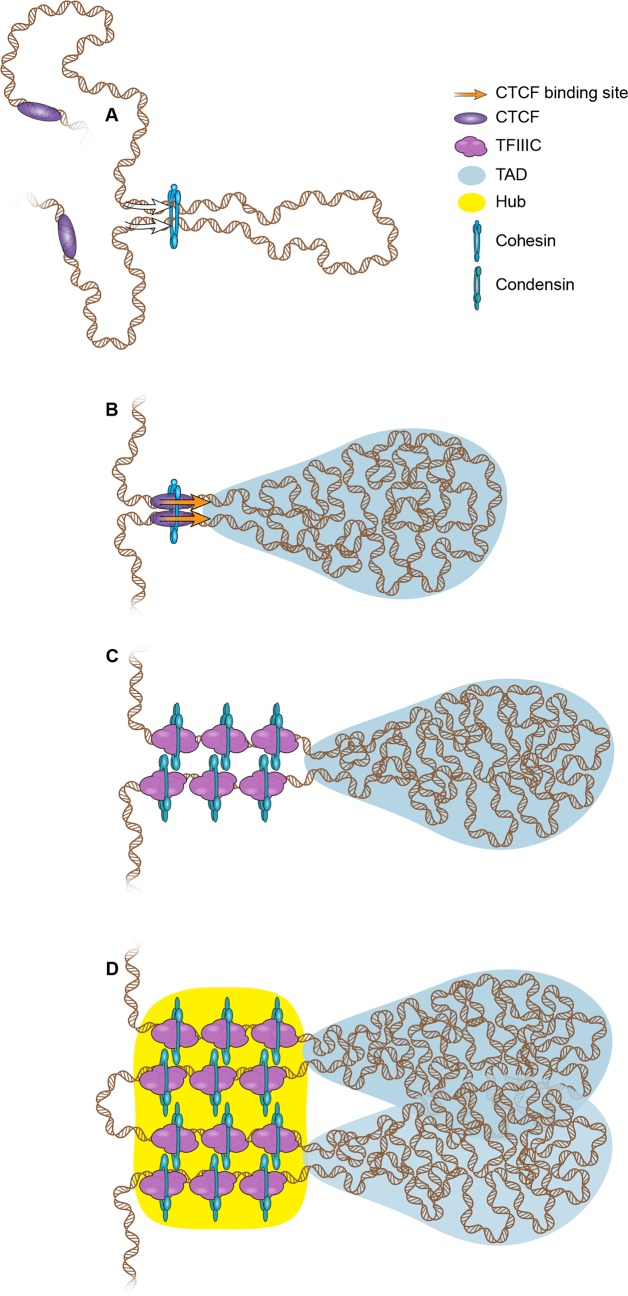
Fig 3. Working models for how adaptor proteins and SMC complexes facilitate the formation of chromosomal domains by loop extrusion.
(A) A loop begins to form as DNA is extruded through the center of an SMC complex ring (blue). Protein rings may move along DNA until an adaptor protein (purple) is encountered that captures the SMC complex ring and halts the extrusion. (B) A region is depicted with cohesin and CTCF facilitating the formation of a topological domain by constraining the sequences at the base. CTCF sites tend to be arranged head-to-head [62]. A single cohesin ring may interact with CTCF and encircle two DNAs to form the base of the loop, which is the boundary between neighboring TADs. Alternatively, two cohesin rings may interact to facilitate the DNA–DNA interaction. (C) Multiple binding sites for condensin II and TFIIIC are found at TAD boundaries. It is not known whether a single condensin ring can encircle two DNAs; we have depicted one ring per DNA, similar to the handcuff model proposed for the bacterial SMC complex in loop extrusion. (D) Interactions between active genes at the base of multiple domains may facilitate the high levels of expression of housekeeping genes, perhaps via a local transcriptional hub (yellow). CTCF, CCCTC-binding factor; SMC, structural maintenance of chromosome; TAD, topological associated domain; TFIIIC, Transcription Factor IIIC.