Abstract
Different symbiotic and pathogenic plant-microbe interactions involve the production of cysteine-rich antimicrobial defensins. In Medicago truncatula, the expression of four MtDefMd genes, encoding arbuscular mycorrhiza-dependent defensins containing an N-terminal signal peptide and exhibiting some differences to non-symbiotic defensins, raised over the time of fungal colonization. Whereas the MtDefMd1 and MtDefMd2 promoters were inactive in cells containing young arbuscules, cells with fully developed arbuscules displayed different levels of promoter activities, indicating an up-regulation towards later stages of arbuscule formation. MtDefMd1 and MtDefMd2 expression was absent or strongly down-regulated in mycorrhized ram1-1 and pt4-2 mutants, known for defects in arbuscule branching or premature arbuscule degeneration, respectively. A ~97% knock-down of MtDefMd1/MtDefMd2 expression did not significantly affect arbuscule size. Although overexpression of MtDefMd1 in arbuscule-containing cells led to an up-regulation of MtRam1, encoding a key transcriptional regulator of arbuscule formation, no morphological changes were evident. Co-localization of an MtDefMd1-mGFP6 fusion with additional, subcellular markers revealed that this defensin is associated with arbuscules in later stages of their life-cycle. MtDefMd1-mGFP6 was detected in cells with older arbuscules about to collapse, and ultimately in vacuolar compartments. Comparisons with mycorrhized roots expressing a tonoplast marker indicated that MtDefMd1 acts during late restructuring processes of arbuscule-containing cells, upon their transition into a post-symbiotic state.
Introduction
The vast majority of terrestrial plants is able to form arbuscular mycorrhizal (AM) symbioses with a group of obligate biotrophic fungi designated Glomeromycota [1, 2]. In return for the supply with hexoses [3] and presumably lipids in the form of palmitic acid [4], AM-fungal mycelia recruit nutrients, especially phosphate, from soils and transport them to the host [5]. It is thought that 450 million years ago, AM fungi facilitated the colonization of land by early plant species [6, 7].
Prior to a colonization of plant roots with AM fungi, strigolactones from the host trigger germination of fungal spores and growth of extraradical hyphae [8, 9]. The hyphae penetrate root epidermal cells via hyphopodia and spread in the cortex from this initial infection site [10, 11]. Both in the epidermis and in the subsequently colonized cells of the root, a pre-penetration apparatus, derived from structures of the cytoskeleton, precedes fungal entry [12]. In cells of the inner cortex, the fungus establishes arbuscules, highly branched, dendritic hyphal ends, being of particular importance for the exchange of nutrients [13]. Nutrient transfer between both partners involves a tight control of the microsymbiont's life-cycle by the host. Fungal arbuscules are regularly degraded after several days, and previously infected cells are restructured for subsequent re-infections [10, 14, 15].
Two AM-specific transcription factors act as key control switches of arbuscule development. With respect to arbuscule build-up, RAM1, together with other GRAS-transcription factors, controls proper arbuscule branching [16]. During the late stages of arbuscule development, the Myb transcription factor MYB1 activates a transcriptional program associated with arbuscule degeneration, leading to the production of different hydrolases and other arbuscule-degrading enzymes [17].
During the formation of active arbuscules, a suite of mycorrhiza-specific phosphate transporter- and other nutrient transporter genes is activated [18, 19]. The transfer of nutrients between fungi and plants throughout the AM symbiosis is mediated by the periarbuscular membrane (PAM), which permanently separates micro- and macrosymbionts [20]. Although the picture of PAM biogenesis, including polarized secretion for spatial expansion of the plasma membrane, is up to now not complete, some key players such as vesicle-associated membrane proteins (VAMPs), syntaxins, vapyrin, and EXO70i were identified [21, 22, 23, 24].
On the other hand, the process of PAM degradation is less clear, and the switch to cell-autonomous arbuscule degeneration is not yet defined, although important components of the degradation program were identified [17]. In this context, Kobae et al. [14] raised the question, why infection units composed of arbuscules in different stages of their life-cycle dissappear in a synchronized manner.
Many eukaryotic cells express genes encoding bioactive, channel-forming amphipathic peptides, such as those belonging to the defensin superfamily [25]. Defensins are not legume-specific, and several members of the defensin family have long been known for their role as defense-related peptides in plants [26]. It was proven that some defensins are translated during the defence reaction towards fungal pathogens [27], while filamentous fungi as well as yeast are considered to be targets as well. Characteristic for plant defensin proteins are an α-helix and a three-stranded, anti-parallel β-sheet, being interconnected by four disulfide bonds that create a cysteine-stabilized αβ motif [28]. In addition, these proteins lack a hydrophobic inner center, leading to a condensed knot-like structure. Some plant defensins have a fifth disulfide bond, which might be important for the stabilization of the protein core, and which allows variation in the surface loops [29]. Finally, the γ-core motif was proposed to be an essential region for the modes of action of the defensin MtDef4 of Medicago truncatula [30]. An insertion into target membranes has been proposed for some plant defensins, since high affinity binding sites exist [31], and since defensins can form weakly anion-selective channels [32].
Defensin genes of Medicago truncatula were found to be expressed in a variety of tissues and in response to different abiotic and biotic stimuli [33]. In the root nodule symbiosis, hundreds of root nodule-specific defensin-like proteins designated nodule cysteine-rich (NCR)-peptides act as effectors of bacteroid development [34, 35]. With respect to the AM symbiosis, Hanks et al. [33] were first to identify defensin genes induced in response to root colonization of Medicago truncatula by the AM-fungus Glomus versiforme, using a combination of transcript sequencing and real-time RT-PCR. Subsequently, several genome-wide transcriptomic studies confirmed that a family of defensin genes is activated during the AM symbiosis of Medicago truncatula [19, 36, 37; 38, 18] and for some, an arbuscule-correlated expression was demonstrated [37, 18].
In congruence with the significant impact of NCR-peptides on terminal bacteroid differentiation in legume root nodules [34, 35], defensins could have important functions for the morphology and life-cycle of arbuscules as well. It is thus the aim of this study to contribute to the functional characterization of mycorrhiza-dependent defensins of M. truncatula, based on a set of four AM-induced MtDefMd genes identified in expression profiling experiments [37, 38, 18]. Focusing on the two MtDefMd genes most strongly activated in a time-course of mycorrhization, we are able to show that their general and cell-specific expression is dependent on key genes of arbuscule formation (MtRam1) and function (MtPt4), respectively. Using translational fusions with subcellular fluorophore markers, we demonstrate that the MtDefMd1 gene is not only specifically transcribed in arbuscule-containing cells, but that the encoded defensin is present during late restructuring processes of arbuscule-containing cells, providing novel insights into their transition into a post-symbiotic state.
Materials and methods
Bioinformatic analyses
Amino acid sequences of mature defensins were aligned using the following settings: Gap open cost: 10.00; gap extension cost: 5; Kyte-Doolittle window lenght 5; polarity logo. RaptorX [39] was used to build a three dimensional model of MtDefMds and AM-unrelated defensins. For this, the three-dimensional structures of HsAFP1 [40], RsAFP1 [41], major pollen allergen Art. v1 [42], and MtDef4 [30] were used as homology models. The coding sequences of MtDefMd1 (Medtr.8g012805.1 in the Medicago truncatula genome [43, 44], Mtr.35854.1.S1_at in the Medicago Gene Expression Atlas (Benedito et al., 2008), MtDefMd2 (Medtr.8g012835.1, Mtr.7210.1.S1_at), MtDefMd3 (Medtr.8g012875, Mtr.3215.1.S1_at), and MtDefMd4 (Medtr.8g012885, Mtr.31214.1.S1_at) were analyzed with SignalP [45], Plant-mPloc [46], and SherLoc2 [47].
Plant cultivars, fungal, and bacterial strains
Medicago truncatula Gaertn. Jemalong A17 (Thierry Huguet, INRA Toulouse, France) was used for all plant experiments. Sterile Rhizophagus irregularis DAOM197198 spores (PremierTech, Rivière-du-Loup, Canada) were applied as fungal inoculum. Escherichia coli DH5α mcr' [48] was used for cloning and the propagation of plasmids. Transgenic roots were induced by Agrobacterium rhizogenes Arqua1 [49].
Cloning of promoter-gusAint fusions
Using genomic DNA of M. truncatula leaves, the promoters of MtDefMd1 and MtDefMd2 (-1 to -1175 bp and -1 to -1457 bp relative to the translational start) were PCR-amplified using primers aaagaattcATTTGTCGTAAATAACCTTGC and aaaaagcttgCTTGCTTAATGTAAATGGAA (MtDefMd1) as well as aaacccgggCGCTTTTAGTTTTCGGTAGAT and aaacccgggGCTTGCTTAATGTAAATGGAA (MtDefMd2). Fragments were cleaved by SmaI and EcoRI/HindIII, respectively, and were cloned into pk18 [50]. The promoters were then subcloned into pGUS-INT [51], containing the gusAint reporter gene. Using SpeI, the transcriptional fusion was cleaved out, Klenow-blunted, and ligated into the SmaI-digested binary vector pRedRoot [52]. Promoter-gusAint fusions cloned in pRedRoot were transferred via electroporation into A. rhizogenes Arqua1.
Cloning of knock-down and overexpression constructs
To obtain an RNAi construct targeting the highly similar MtDefMd1 and MtDefMd2 genes, the coding sequence of MtDefMd1 was PCR-amplified from position +2. attB sites were added for cloning into the entry vector pDONRTM221 (Gateway®-System, Invitrogen, Karlsruhe, Germany), using the BP clonase reaction. The LR clonase reaction was used for cloning into the destination vector pK7GWIWG2(II)-Q10:DsRED [53].
To obtain an overexpression construct, the coding sequence of MtDefMd1 (656 bp, containing introns) was PCR-amplified, cleaved with BamHI and SpeI and ligated into vector p9RFP [54, 55] containing the MtPt4 or ubiquitin promoters, respectively. SpeI and BamHI recognition sites as well as 5 additional adenosin bases were added to the primers aaaaactagtATGGCTTCCTCTGCTCTTAAAT and aaaaggatccTTAGCAGTTGAAGTAACAGAAGCAAG.
Cloning of constructs for subcellular localizations
A cassette of the native promoter and the coding sequence of MtDefMd1 (-1 to -1175 bp; 656 bp coding region) was cloned into vector p35S-OLI-mGFP6 [56]. This vector encodes an N-terminal GGRSPGGS oligopeptide (OLI) extension of mGFP6 that serves as a flexible linker to the protein of interest. The following cloning strategy were used: SphI and KpnI recognition sites were added to the primers for cloning the MtDefMd1 promoter (aaagcatgcATTTGTCGTAAATAACCTTGCCT/ aaaggtaccGCTTGCTTAATGTAAATGGAAATG), and KpnI and BglII were used for the coding sequence (aaaggtaccaaaaATGGCTTCCTCTGCTCTTA/aaaagatcttcctccGCAGTTGAAGTAACAGAAG), allowing the translation of an MtDefMd1-OLI-mGFP6 fusion protein under the control of the native MtDefMd1 promoter instead of p35S. All primers were extended by 3 additional adenosin bases. The MtDefMd1-OLI-mGFP6 fusion was released using EcoRI and HindIII. The fragment was blunted using Klenow polymerase and ligated into the blunted SalI site of binary vector pBIN:ER-ck [57] expressing an ER-CFP marker. Finally, the fusion of the MtBcp1 promoter and signal peptide region to the coding sequence of mCherry (MtBcp1SP-mCherry) was released from pCMbB-TMEr [58] using Ecl136II and SmaI. The insert was introduced into the SmaI site of the vector containing the MtDefMd1-OLI-mGFP6 and ER-CFP fusions, resulting in a three-fluorophore vector designated pBIN:ER-ck:MtDefMd1-OLI-mGFP6:MtBcp1SP-mCherry. Vector pBIN:tp-gk [57] was used for tonoplast localization.
Cultivation of M. truncatula plants
Plants were cultivated in a phytocabinet (Klimaschrank KPS 1700, Weisshaar, Bad Salzuflen, Germany) with 16 h/d light (Osram FLUORA L 18WI77 light tubes) at 22°C with a relative humidity of 60%. Plants were fertilized with ½ strength Hoagland's solution [59] containing 20 μM phosphate. The solution was prepared with deionised water; the pH was adjusted to 6.3 with H2SO4.
Induction of transgenic M. truncatula roots and inoculation of M. truncatula with AM fungi
Transgenic M. truncatula roots were generated as described previously [60]. In short, Agrobacterium rhizogenes Arqua1 strains were grown for two days at 30°C on selective TY (0.5 g/l tryptone; 0.3 g/l yeast extract; 0.07 g/l CaCl2x2H2O) agar plates. Cells were resuspended in 4 ml PS buffer (40 mM Na2HPO4x2H2O, 85 mM NaCl, 17 mM KH2PO4; pH 7 adjusted with HCl). M. truncatula seedlings were moisturized and cleared of their testa. The bacterial solution was injected in the hypocotyl of the seedlings with a sterile syringe. Subsequently, seedlings were planted in sterile seramis® (Seramis GmbH, Mogendorf, Germany) and incubated over night at 18°C in the dark. Finally, they were transferred to the phytocabinet climate chamber. Transgenic hairy roots were screened and mycorrhized four to five weeks after germination. For most experiments, sterile R. irregularis spores were used for the inoculation of M. truncatula wild type and transgenic roots. 2000 spores were incubated with the shaded plant root for three hours in liquid. Subsequently, plants were potted in 9×9 cm pots with seramis (Seramis GmbH, Mogendorf, Germany) as a substrate. Remaining spores were pipetted on the roots. For RNAi experiments, R. irregularis inoculum from a leech preculture was used (Bettina Hause, IPB, Halle, Germany).
Isolation of plant RNA from M. truncatula roots
RNA isolations were carried out using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). ß-mercaptoethanol was added to the RLT buffer and 600 μl of this mix were added to each sample. Tissues were disrupted using the FastPrep®-24 (MP Biomedicals, Santa Ana, USA) for 5 times 30 s (6.5 m/s). The RNA concentrations were measured using a Nanodrop (Thermo Fisher Scientific, Langenselbold, Germany) and checked on agarose gels.
Real-time RT-PCR
Real-time RT-PCR analyses were performed using the SensiFAST™ SYBR® No-ROX One-Step Kit (Qiagen, Hilden, Germany) with primers listed in the S1 Table. Primers were designed to match an annealing temperatures of 55°C and were tested for gene-specific amplifications. 5 ng of total RNA were used as a template in 20 μl for the mycorrhization time course, the ram1-1 and the MtDefMd1 overexpression experiment. Otherwise, 50 ng of total RNA were used. RT-PCR reactions followed a three-step cycling program: reverse transcription at 45°C for 10 min; polymerase activation at 95°C for 2 min; PCR amplification with 40 cycles at 95°C for 5 sec, 55°C for 10 sec, and 72°C for 8 sec.
For all genes, three technical replicates for each of 12 individual transgenic roots—if not stated differently—were measured. The housekeeping gene MtTefα was used for normalization. To relate gene expression to the degree of fungal colonization or the presence of active arbuscules, a ratio with GiTubα or MtPt4 expression was calculated, respectively. The degree of fungal colonization was estimated by measuring the expression of the GiTubα gene encoding an alpha-tubulin [37], whereas the MtPt4 transcript amount served as an estimate for the presence of active, phosphate-transporting arbuscules [61]. The mean and standard error of the mean of all biological replicates was calculated after normalization and was visualized, if not stated differently. Using MS Excel, two-tailed Student t-tests were performed to check differences in gene expression between mutant-, RNAi-, overexpression-, and wild type roots. To determine, if the expression of AM-related defensin and AM marker genes is congruent during the course of mycorrhization, Pearson correlation was calculated by using the MS Excel KORREL function for comparing the average gene expression levels at each time point.
Histological studies
To study the activity of promoter-gusAint fusions, transgenic roots were incubated in GUS staining buffer described previously [51]. Cells with the strongest promoter activity were visible after 3 hours. After overnight staining, also cells with a weak promoter activity were stained. Finally, roots were rinsed with water.
To stain fungal structures for quantification of AM fungal matter via the gridline intersection method [62] and confocal microscopy, 1–2 cm root sections were incubated in 10% (w/v) KOH at 95°C for 7 minutes. Consecutively, the roots were washed three times with water and incubated in staining solution with 20 μg/ml Alexa-WGA Fluor 488 in 1x PBS (0.14 M NaCl, 2.7 mM KCl, 1 mM Na2HPO4x2H2O, 1.8 mM KH2PO4; pH 7.3) for 12–24 hours. Samples were protected from light. Finally, excessive dye was washed out with water. Mycorrhized root samples were randomly collected and pooled from each plant. Three pools of 6 root sections were photo-documented by confocal microscopy.
Detection of reporter proteins via fluorescence microscopy
Transgenic M. truncatula roots were identified using a stereo microscope (Leica MZ10F, Sohns, Germany). 1–2 cm sections of mycorrhized roots were selected. For the localization of fluorescent reporter proteins, longitudinal root sections were cut by hand with a razor blade. These sections were transferred into a physiological buffer (39 mM Na2HPO4x2H2O, 86 mM NaCl, 22 mM KH2PO4; pH 7). GFP (500–520 nm), CFP (460–490 nm) and mCherry (599–621 nm) were detected in the inner root cortex using a hybrid detector and confocal microscopy (Leica TCS SP8 MP, Sohns, Germany). To evaluate, if the detected signals originate from the correct fluorophore, lambda-scans were performed in the range of 498–553 nm (mGFP6), 458–513 nm (CFP), and 575–635 nm (mCherry). Each lambda-scan contained eleven detection steps with 5 (mGFP6 and CFP), and 11 (mCherry) nm bandwidth, respectively. For promoter-GUS studies, an Eclipse TE2000-E inverse confocal laser scanning microscope (Nikon GmbH, Düsseldorf, Germany) and software EZ-C1 (Nikon GmbH, Düsseldorf, Germany) were used.
Results
Arbuscular mycorrhiza-related defensins differ from defence-related defensins
The four AM-related defensin genes MtDefMd1, MtDefMd2, MtDefMd3, and MtDefMd4 (identifiers Mtr.35854.1.S1_at, Mtr.7210.1.S1_at, Mtr.3215.1.S1_at, and Mtr.31214.1.S1_at in the Medicago Gene Expression Atlas [63], respectively) were primarily identified based on their strong up-regulation in mycorrhizal roots [37].
In the deduced amino acid sequences of MtDefMd1-4, signal peptides were located up to the 29th amino acid (Fig 1, S2 Table), suggesting that the defensins MtDefMd1-4 are secreted. Since it is known that the ER-Golgi network is redirected during the establishment of arbuscules [64], a targeting of MtDefMd1-4 to the periarbuscular space (PAS) in symbiotic cells is thus possible.
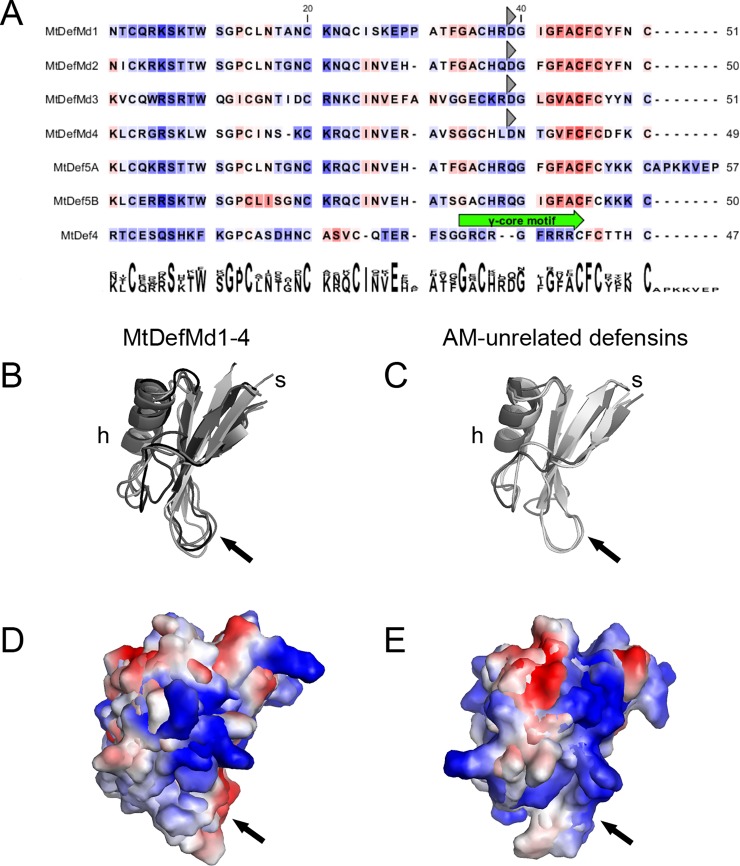
Fig 1. Sequence analyses of of AM-dependent defensins MtDefMd1-4 and AM-unrelated defensins.
Secondary structures of MtDefMd1-4 and AM-unrelated defensin-like proteins of M. truncatula (A), a representation of their three-dimensional structures (B and C), as well as surface electrostatics (D and E) are shown. Predicted signal peptides were removed from the mature amino acid sequences. Consecutively, the defensins were aligned based on their secondary structures. Background colorisation of the amino acids (in A) indicate hydrophobicity in a scale from red to blue (red: high hydrophobicity). A conserved aspartic acid in the C-terminal region of MtDefMds is marked with a grey triangel. For the bi-domain defensin MtDef5, the domains MtDef5A (including a 7 amino acid linker towards the MtDef5B domain) and MtDef5B are shown. After modelling the three-dimensional structures of the MtDefMd1-4, MtDef4 and MtDef5A/B defensins, they were visualized (B and C) and their surface electrostatics were calculated (D and E). The region congruent to the γ-core motif is indicated with arrows. The following proteins were used for comparisons in addition to MtDefMd1-4: MtDef5 A [65], MtDef5 B [65]and MtDef4 [30].
To investigate the secondary structures of the AM-dependent defensins MtDefMd1-4 in comparison to the AM-unrelated defensins MtDef4 [30] and MtDef5A/B [65], the amino acid sequences of the mature proteins were compared (Fig 1A). Subsequently, the three-dimensional structures of MtDefMd1-4 [based on HsAFP1, 40; RsAFP1, 41; HsAFP1, 40; and major pollen allergen Art. v1, 42], MtDef5A [based on HsAFP1, 40], MtDef5B [based on HsAFP1, 40], and MtDef4 [MtDef4, 30] were modeled and visualized (Fig 1B and 1C).
AM-dependent MtDefMds as well as AM-unrelated defensins are structurally conserved, being composed of an α-helix, three β-strands and the γ-core motif (Fig 1B and 1C). The four AM-related defensins are characterized by a hydrophobic C-terminus and an acidic amino acid (aspartic acid) at position 42 (Fig 1A). Although this discriminates them from the AM-unrelated defensin MtDef4 [30], both domains of the AM-unrelated defensin MtDef5A/B, which was shown to confer a broad antifungal activity [65], also display hydrophobic amino acids at the C-terminus (Fig 1A). Interestingly, some areas of the MtDefMd1-4 and AM-unrelated defensins exhibit a different distribution of cationic amino acids (Fig 1D and 1E) and the region corresponding to the γ-core motif, a variable loop essential for entry of MtDef4 in fungal cells [30], displays a differing pattern of surface electrostatics in AM-dependent and -unrelated defensins (Fig 1D and 1E).
In conclusion, MtDefMd proteins exhibit some differences to non-symbiotic defensins and are predicted to be secreted, suggesting a function at the interface between plants and AM fungi.
MtDefMd transcription is upregulated in the course of mycorrhization
A mycorrhization time course was set to define the temporal regulation of MtDefMd1-4 expression. M. truncatula plants were inoculated with R. irregularis spores using ½ strength Hoagland's solution containing 20 μM phosphate for fertilization, and the root system was harvested from 7 to 42 days post inoculation (dpi). The expression of MtDefMd1-4, a fungal α-tubulin (GiTubα) gene [36], and two M. truncatula AM marker genes (MtPt4 [66], MtMyb1 [17]) was measured via real-time RT-PCR (Fig 2), using the MtTefα gene encoding a translation elongation factor for normalization.
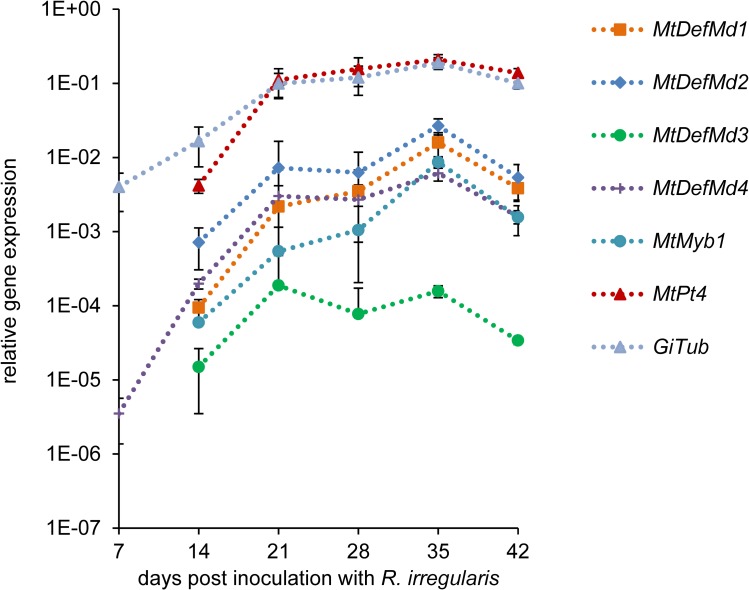
Fig 2. Relative expression of MtDefMd1-4 and selected AM marker genes in M. truncatula roots in a time course of mycorrhization.
Expression of MtDefMd1-4, the fungal α-tubulin gene GiTubα [36] as well as the M. truncatula AM marker genes MtPt4 [66] and MtMyb1 [17] is given in relation to the expression of MtTefα. Plant roots were harvested weekly from 7 to 42 days post inoculation with R. irregularis. Biological replicates were three pools of four plant roots per treatment. The standard deviation of the three biological replicates is given.
MtDefMd1, MtDefMd2, and MtDefMd4 expression strongly correlated (r≥0.8) to that of all AM marker genes, while the correlation of MtDefMd3 transcription was lower but still positive (r≥0.51, S3 Table). Transcription of the MtDefMd genes thus followed the colonization of plant roots with R. irregularis, estimated by the expression of the fungal and plant AM marker genes GiTubα, MtPt4, and MtMyb1 (Fig 2). Whereas transcription of the fungal α-tubulin gene and MtDefMd4 already rose from 7 dpi in a constant manner (Fig 2), expression of the other three MtDefMd genes as well as the AM markers genes MtPt4 [66] and MtMyb1 [17] strongly enhanced from 14 dpi on. It can be concluded that fungal mass enhanced from the start of inoculation, while arbuscules were effectively built from 14 dpi. Interestingly, at 42 dpi, the expression of all genes measured decreased, possibly due to a control of the degree of mycorrhization by the plant.
We conclude that on a whole-root level, the expression of the four AM-activated MtDefMd genes correlates with the expression of marker genes for root colonization and AM formation.
MtDefMd1 and MtDefMd2 promoters are active in cells with fully developed arbuscules
Since MtDefMd1 and MtDefMd2 are the defensin genes activated most strongly during the later colonization stages of M. truncatula roots with R. irregularis (Fig 2), we focused on these genes for further studies.Transcriptional fusions of the promoters of MtDefMd1 and MtDefMd2 with the gusAint reporter gene were expressed in transgenic roots of M. truncatula A17 wild type to investigate their spacio-temporal expression during AM. Roots were mycorrhized for 18 dpi and analyzed via GUS and Alexa-WGA Fluor 488 stainings (Fig 3).
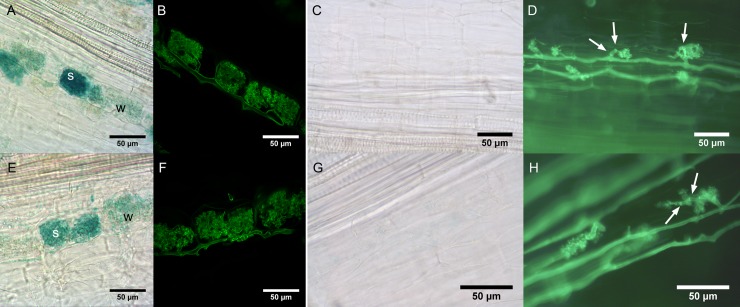
Fig 3. Histochemical localization of MtDefMd1 and MtDefMd2 promoter activities.
Activities of MtDefMd1 (A-D) and MtDefMd2 (E-H) promoters were studied in transgenic, mycorrhized roots of M. truncatula A17 wild type (A, B, E, and F) and pt4-2 roots (C, D, G, and H). Representative images of roots after 18 (A, B, E, and F) or 56 (C, D, G, and H) days post inoculation with R. irregularis. The GUS-stainings (A, C, E, and G) as well as the Alexa-WGA Fluor 488 stainings (B, D, F, and H) were performed over night. Septa are denoted by arrows. Abbreviations: w, cells with weak promoter activity; s, cells with strong promoter activity.
Activities of the MtDefMd1 and MtDefMd2 promoters were investigated from 7 to 56 dpi. In wild type roots, their activities varied from cell to cell, as shown for 18 dpi in (Fig 3A and 3E). While in pre-arbuscular stages and in cells with arbuscules appearing very young, no promoter activities were detected, GUS staining was present in cells with fully developed and apparently active arbuscules (Fig 3A and 3E). This indicates that the MtDefMd1 and MtDefMd2 promoters are activated in cells containing mature arbuscules.
To correlate MtDefMd1 and MtDefMd2 promoter activities with arbuscule development, the corresponding gusAint fusions were expressed in pt4-2 mutants displaying premature arbuscule degeneration (PAD) [67]. Interestingly, there was no activity of the MtDefMd1 and MtDefMd2 promoters detectable even after 56 dpi (Fig 3C, 3D, 3G and 3H) and overnight staining. This observation corresponds to a downregulation of these genes in pt4-2 mutant in comparison to wild type roots [17].
We conclude that while MtDefMd1 and MtDefMd2 promoter activities are correlated with the presence of fully developed, active arbuscules, pre-mature arbuscule degradation does not involve transcription of these defensin genes. This suggests that MtDefMd defensins are not part of the PAD program activated in pt4-2 mutants.
MtDefMd1 and MtDefMd2 expression is impaired in ram1-1 mutants forming bird's-feet arbuscules
To pinpoint the stage of arbuscule development, which coincides with MtDefMd1 and MtDefMd2 transcription, ram1-1 mutants [68] were used. These mutants lack the trancription factor MtRam1, an important regulator that coordinates the further development of arbuscules post the bird's-feet stage by activating the transcription of a suite of AM-associated genes. To study MtDefMd1 and MtDefMd2 expression in relation to arbuscule maturation, mycorrhized ram1-1 and wild type RNA samples were analyzed via real-time RT-PCR (Fig 4).
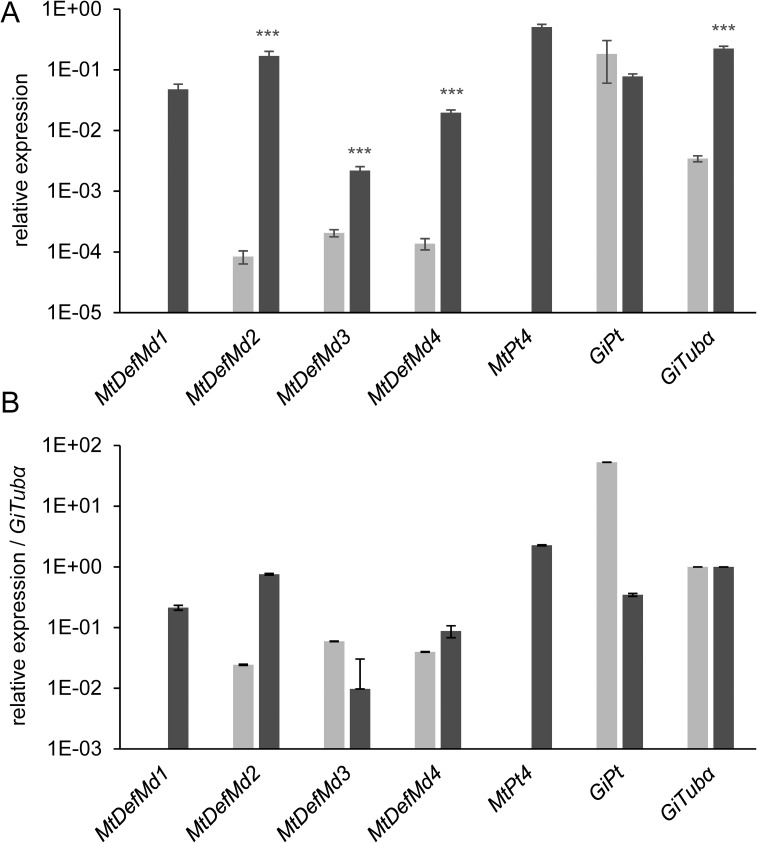
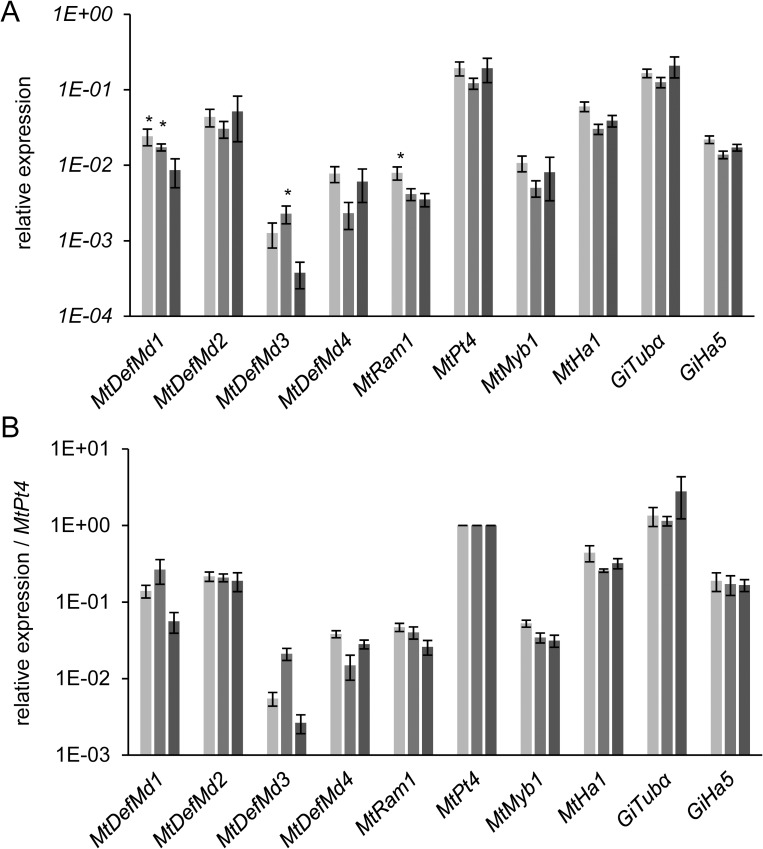
Fig 4. Relative expression of MtDefMd and selected AM-marker genes in mycorrhized M. truncatula A17 wild type and ram1-1 roots.
Transcript amounts are shown relative to MtTefα (A) and were additionally normalized by building a ratio with GiTubα-expression (B). ram1-1 measurements are colored in light grey, the corresponding wild type measurements in dark grey. Roots were harvested at 36 days post inoculation with R. irregularis. n = 12 biological replicates, depicted is the standard error of the mean. Statistical significance: *** p≤0.0005.
Whereas no MtDefMd1 and MtPt4 transcripts were detectable in mycorrhized ram1-1 mutants, the expression of MtDefMd2, MtDefMd3, MtDefMd4 and GiTubα appeared strongly reduced. Only a fungal phosphate transporter gene (GiPt) was not affected in expression. When normalizing gene expression to the level of root colonization by building a ratio with GiTubα expression, only the expression of MtDefMd1, MtDefMd2, and MtPt4 appeared absent or strongly downregulated in ram1-1 mutants (Fig 4B).
Based on these studies, the time point of MtDefMd1 and MtDefMd2 action can be placed in the arbuscule-containing cells, post the bird's-feet stage. This corresponds to our observation that the promoters of AM-related defensin genes are active in cells containing fully developed arbuscules.
Knock-down of MtDefMd1 and MtDefMd2 does not affect expression of AM marker genes
Since MtDefMd1 and MtDefMd2 promoter activities were connected to fully developed arbuscules, we investigated, if their knock-down would affect arbuscule development. To reduce transcription of MtDefMd1 and MtDefMd2 simultaneously, an RNA interference (RNAi) construct targeting both genes was expressed in mycorrhized transgenic M. truncatula roots.
Due to high similarities between the short MtDefMd1 and MtDefMd2 sequences, it was not possible to design gene-specific primers outside the region affected by the RNAi construct. Therefore, both genes were measured simultaneously in mycorrhized transgenic RNAi- and control roots (Fig 5). Since MtDefMd2 expression alone was knocked down to 5.9%, the concomitant MtDefMd1 and MtDefMd2 reduction to appr. 3% indicated that similar to MtDefMd2, only residual amounts of MtDefMd1 transcripts remained, confirming an efficient knock-down of both MtDefMd1 and MtDefMd2 expression (Fig 5A). In contrast, transcript amounts of MtDefMd3, MtDefMd4, MtPt4, and GiTubα did not differ significantly between RNAi- and control roots (Fig 5A and 5B). When normalized to the number of active arbuscules by building a ratio to MtPt4-expression (Fig 5B), the reduction of MtDefMd1 and MtDefMd2 transcript amounts remained very strong.
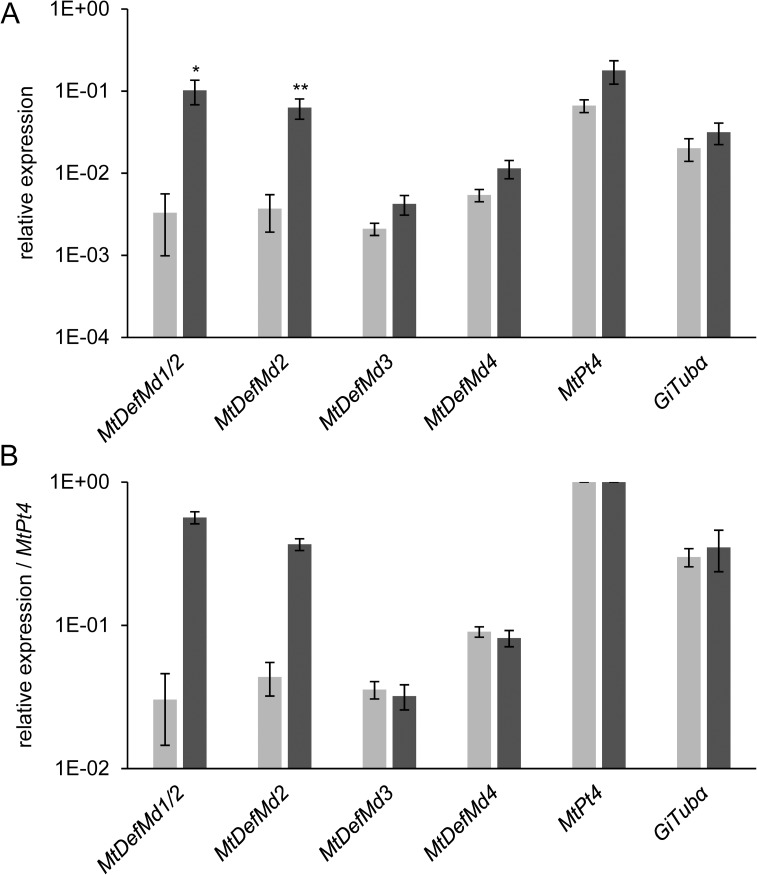
Fig 5. Relative expression of MtDefMd and selected AM marker genes in mycorrhized RNAi:MtDefMd1/2 and RNAi:gusAint transgenic control roots of M. truncatula.
Transcript amounts are shown relative to MtTefα (A) and were additionally normalized by building a ratio to MtPt4-expression (B). Measurements from RNAi:MtDefMd1/2 roots are colored in light grey, corresponding RNAi:gusAint control measurements in dark grey. Roots were harvested at 28 days post inoculation with R. irregularis. n = 12 biological replicates, depicted is the standard error of the mean. Statistical significances: * p≤0.05, ** p≤0.005.
Since the severe knock-down of MtDefMd1 and MtDefMd2 did not lead to significant changes in MtPt4 and GiTubα expression, we conclude that these MtDefMd genes are not essential predecessors for a successful fungal colonization and for the formation of active, phosphate-transporting arbuscules.
Overexpression of MtDefMd1 activates MtDefMd3 and MtRam1
Due to the possibility that the four AM-related MtDefMds studied here and other yet undiscovered AM-related defensins act in parallel, it is difficult to elucidate their function by knocking down two of them. We thus decided to get insights into the role of defensins during AM by overexpressing the MtDefMd1 gene in mycorrhized transgenic roots under the control of the Ubiquitin- and the MtPt4-promoter, mediating a general or a specific expression in arbuscule-containing cells, respectively (Fig 6).
Fig 6. Relative expression of MtDefMd1 and selected AM marker genes in mycorrhized MtDefMd1-overexpression and pPT4:gusAint-expressing transgenic control roots of M. truncatula.
Transcript amounts are shown relative to MtTefα (A) and were additionally normalized by building a ratio to MtPt4-expression (B). Measurements of pPT4:MtDefMd1 overexpression roots are coloured in light grey, measurements of pUbi:MtDefMd1 overexpression roots in medium grey, and measurements of pPT4:gusAint control roots in dark grey. Roots were harvested at 28 days post inoculation with R. irregularis. n = 12 biological replicates, depicted is the standard error of the mean. Statistical significance: * p≤0.05.
Overexpression of MtDefMd1 under control of the MtPt4- and the Ubiquitin-promoter led to a significant 2.8-fold and 2-fold overexpression of this defensin, respectively (Fig 6A). Whereas MtDefMd3 was co-activated by MtDefMd1 overexpression controlled by the Ubiquitin-promoter, expression of MtDefMd2 and MtDefMd4 was not affected (Fig 6A and 6B). Interestingly, transcription of MtRam1 was 2.3-fold upregulated in case of an MtDefMd1 overexpression driven by the MtPt4 promoter. To further assess co-activation of MtRam1 and MtDefMd3, transcript levels were normalized by building a ratio to MtPt4 expression (Fig 6B), since the MtDefMd1 and MtDefMd2 promoters are specifically activated in the arbuscule-containing cells. Thereby, the overexpression of MtDefMd1, MtRam1 and MtDefMd3 was verified in relation to the formation of active arbuscules (Fig 6A and 6B).
Knock-down of MtDefMd1/2 transcription and overexpression of MtDefMd1 has no significant impact on arbuscule size
To investigate, whether an MtDefMd1/2 knock-down or the overexpression of MtDefMd1 influences fungal colonization, phenotypical studies were performed in comparison to control roots. Due to the fact that on a whole-root level, no significant changes in the frequency of mycorrhization or the formation of arbuscles were observed (S4 Table, S5 Table), the distribution of arbuscule sizes was assessed via confocal microscopy of Alexa-WGA Fluor 488-stained root sections of mycorrhized RNAi:MtDefMd1/2 and MtDefMd1-overexpression in relation to pPt4:gusAint control roots (Fig 7).
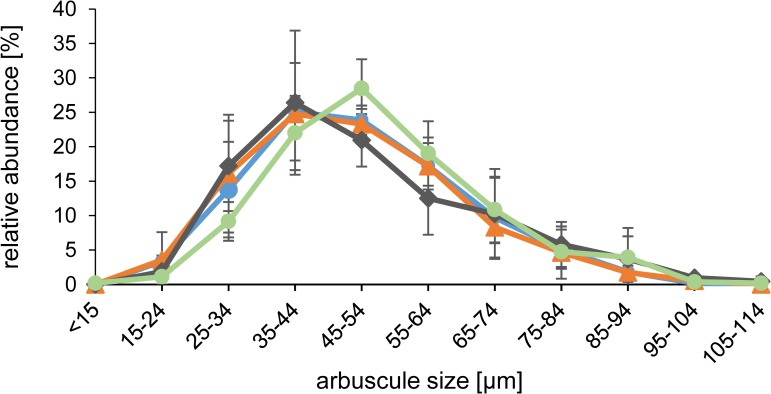
Fig 7. Size distribution of arbuscules in mycorrhized M. truncatula MtDefMd1-overexpression, MtDefMd1/2-knock-down, and pPT4:gusAint controls roots.
Arbuscules were sorted into one of eleven size categories. In total, the size distribution of arbuscules in pUbi:MtDefMd1-overexpression (647 arbuscules), pPt4:MtDefMd1-overexpression (509 arbuscules), RNAi:MtDefMd1/2-knock-down (625 arbuscules), and pPt4:gusAint control roots (529 arbuscules) is depicted in orange, blue, green, and grey, respectively. Roots were harvested at 28 days post inoculation with R. irregularis. For each construct, three pools of root fragments, each pool being derived from four plants, were analysed. Depicted is the standard error of the mean.
Although the transcription factor gene MtRam1 was upregulated by MtDefMd1 overexpression (Fig 6), the sorting of arbuscules in different size categories indicated that similar to the whole-root level, there is no significant difference to the control roots (Fig 7). Thus, the global distribution of arbuscule sizes was congruent in MtDefMd1 overexpression roots. Similarly, although a slight shift towards larger sizes was evident (Fig 7), no significant alteration of arbuscule sizes was recorded for the RNAi:MtDefMd1/2 knock-down roots.
We conclude that neither an appr. 97% reduction of MtDefMd1/2 transcription nor an up to 2.8-fold overexpression of MtDefMd1 has a detectable effect on the steady-state distribution of arbuscule sizes.
MtDefMd1 acts during late restructuring stages of arbuscule-containing cells
From our promoter studies, we concluded that the two highly similar MtDefMd1 and MtDefMd2 genes are expressed in cells containing fully developed arbuscules. To monitor MtDefMd1 distribution and accumulation during the life-cycle of an arbuscule-containing cell, we designed a triple fluorophore reporter construct. First, we fused the MtDefMd1 coding region to mGFP6. In the resulting MtDefMd1-mGFP6 fusion, both regions are separated by a flexible linker. This modification enables free rotation between the two regions and was previously used to localize symbiosome-targeted proteins in the infected cells of root nodules [56]. To achieve a correctly regulated expression in the arbuscule-containing cells, the MtDefMd1-mGFP6 fusion was expressed under the control of the MtDefMd1 promoter. To visualize protein secretion that is known to be transiently redirected towards the arbuscules [64], we introduced a constitutive endoplasmatic reticulum (ER) CFP-marker [57]. The signal peptide of MtBcp1 fused to mCherry [58] and expressed under the control of the MtBcp1 [19] promoter, was used to define consecutive stages of the arbuscule life-cycle. MtBcp1 is present in the stem region of young and is more evenly distributed in the PAM of mature, fully developed arbuscules, thus allowing to define arbuscule stages [58]. All reporter fusions were concomitantly expressed in mycorrhized transgenic roots, and confocal microscopy was used to locate mCherry, CFP, and GFP fluorescence in the inner cortex (Fig 8).
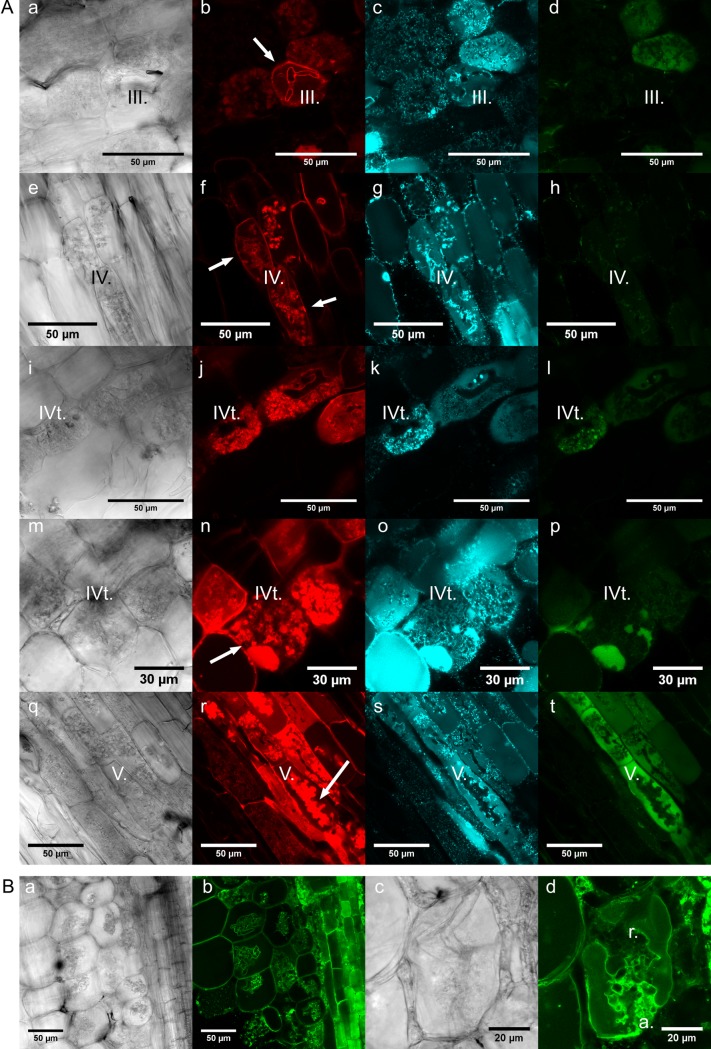
Fig 8. Localization of MtDefMd1-mGFP6 and additional fluorescence marker proteins in mycorrhized M. truncatula roots.
Differential interference contrast micrographs of razor blade hand-cut sections of transgenic M. truncatula roots are shown (A; a, e, i, m, and q; B; a and c). Confocal microscopy was used to localize a fusion of the signal peptide of MtBcp1 with mCherry under the control of the MtBcp1 promoter (A; b, f, j, n, and r), an ER-CFP fusion under the control of the 2x35S-promoter (A; c, g, k, o, and s), and an MtDefMd1-mGFP6 fusion under the control of the MtDefMd1 promoter (A; d, h, i, p, and t). Additionally, a tonoplast membrane-directed GFP fusion under the control of a 2x35S-promoter was studied (B; b and d). Roots were mycorrhized with R. irregularis for six weeks. Arrows in the MtBcp1SP-mCherry micrographs indicate structures referred to in the text. Abbreviations: a., arbuscule branches; r., restructuring; III, bird's-feet arbuscule; IV, active arbuscule; IVt, fully developed arbuscule prior to collapsing; V, collapsed arbuscule.
In congruence to the MtBcp1-localisation reported by Ivanov and Harrison [58], the trunks of arbuscules in the bird's-feet stage (stage III.) were positive for MtBcp1SP-mCherry (Fig 8A, b). In addition, also the symplast membrane of cells in the symbiotic root tissues exhibited a certain level of mCherry fluorescence (Fig 8A, f). In mature, fully developed arbuscules, presumably active in metabolite exchange (stage IV.), MtBcp1SP-mCherry signals expanded to the whole PAM (Fig 8A, f and j).
Since the ER-marker was originally designed for the analysis of cell organelles in Arabidopsis thaliana [57], its functionality in M. truncatula had to be verified. It was expected that a spider web structure surrounding the nucleus, granular bodies at the edges of the cells as well as components of the cytoskeleton used to direct transport vesicles were stained. However, this was only the case for cells not containing arbuscules (Panel A in S1 Fig). In arbuscule-containing cells of the inner cortex, a close connection of ER-structures to the PAM (Fig 8A, c, g, k, o and s) and a CFP signal in the vacuole of GFP-positive cells (Fig 8A, o) was detected, indicating a massive reorganisation of vesicle traffic to or from the PAM.
MtDefMd1-GFP signals were only observed in a fraction of the arbuscule-containing cells (Fig 8A, I, p and t) and were not detected in cells prior to arbuscule branching (Fig 8A, d). In cases of low GFP signals, small dots and spheres in close proximity to the PAM of fully developed arbuscules were apparent (Fig 8A, l). These signals match the patterns of CFP-labeled ER-structures (Fig 8A, k, o and s) and while they are too large for being vesicles, they might be conglomerates of such. This points towards MtDefMd1 being part of the transportome redirected to the PAM. The MtDefMd1-mGFP6 signal increased strongly in cells with older, collapsing arbuscules (Fig 8A, p and t). These were characterized by structures appearing condensed and clumpy. In those cells, the mGFP6 signal gets very intensive and seems to be more and more present in the vacuole (Fig 8A, p and t).
To further investigate if the GFP pattern of intensively coloured cells matches restructuring processes of the vacuole during the AM symbiosis, transgenic roots expressing a tonoplast membrane GFP-marker [57], were mycorrhized and GFP-fluorescence was analyzed (Fig 8B, b and d). Whereas in non-mycorrhized roots, the space occupied by the cytoplasm, i.e. the volume close to the cell wall and cytoplasmic transvacuolar strains, are indirectly visualized (Panel B in S1 Fig), the space taken by the arbuscule is clearly defined by the surrounding GFP-labeled tonoplast (Fig 8B, b and d). The remaining space comprises the cytoplasm, the PAS and the symbiotic membranes from both organisms. Tonoplast membranes switch from a very close contact with arbuscule branches to a more loose one (Fig 8B, d, areas a and r). The morphology of cells exhibiting this phenomenon fits to those containing arbuscules in late stages of their life-cycle, characterized by a high level of MtDefMd1-mGFP6 (Fig 8A, p).
Our MtBcp1SP-mCherry, ER-CFP, and MtDefMd1-mGFP6 colocalization indicates that arbuscule structures undergo a transitional change between stage IV. (fully developed) and V. (degrading), which will be termed IVt. from now on. Once the distinct MtBcp1-mCherry signal in the arbuscule trunk (stage III.) extends towards the tips of the arbuscule branches (Fig 8A, j), the MtDefMd-mGFP6 signal starts to co-localize to these structures (Fig 8A, l; stage IV.t). Now, the distal tips of the arbuscule branches tend to get crescent-shaped or swollen, collapse (Fig 8A, j and n), and the space occupied by the arbuscule looks increasingly clumpy. Occasionally, the cell lumen contains non-fluorescent spheres of differing sizes, possibly representing partially degraded arbuscules (Panel A in S1 Fig). Strongly mGFP6-positive, most likely vacuolar compartments are increasingly created during stage IVt. and are abundant in stage V. (Fig 8A, p and t). The creation of fluorophore-positive vacuoles indicates a recycling of PAM-targeted vesicles and proteins, which were previously located in the perisymbiotic membrane. Since the increase of MtDefMd1-mGFP6 levels is strongest during this rather short and late stage of the arbuscule life-cycle (Fig 8A, l), it marks the beginning of the turnover from a symbiotic into a post-symbiotic cell.
Discussion
AM-related defensins of the MtDefMd family can be distinguished from other antimicrobial peptides. First, the γ-core motif, which was vital for MtDef4 entry into Fusarium graminearium [30], is strikingly different in AM-related defensins. Second, a calculation of surface hydrophobicities identified a distinct pattern of hydrophobic amino acids and an aspartic acid residue close to the C-terminal region only for AM-related defensins, suggesting specific symbiotic targets for their function.
The shift of MtBcp1-mCherry from arbuscule trunks to the PAM allowed us not only to differentiate between pre-mature, fully developed, and collapsing arbuscules, but also to relate MtDefMd1-mGFP6 signals to these stages of arbuscule formation and degradation (Fig 8A, h to t). In contrast to MtBcp1-mCherry, MtDefMd1-mGFP6 signals were not detected prior to arbuscule branching (Fig 8A, d). Together with our observation that the strongest MtDefMd1-mGFP6 signals were found in cells with collapsing arbuscules and increasingly in vacuoles (Fig 8A, p and t), this indicates that MtDefMd1 functions during late stages of the arbuscule life-cycle, and here during the initiation of a regular turnover of symbiotic structures that involves massive restructuring of the PAM (Fig 8A, transition from h to l, l to p, and p to t). This is in line with our observation that the MtDefMd1 and MtDefMd2 promoters are activated in fully developed arbuscules and that the premature arbuscule degradation observed in pt4-2 mutants does not involve MtDefMd genes.
Using in planta time-lapse imaging, it was shown that in contrast to their formation, arbuscule collapse is a rapid event [14]. Recent evidence indicates that the arbuscule degeneration program activated by MtMyb1 [17] plays an important role to initiate this step. In this light, it is conclusive that MtDefMd1 accumulation happens in a short period of time, defined by the IVt. stage (Fig 8A, i to p). The strong RNAi-mediated knock-down of MtDefMd1 and MtDefMd2 had no detectable impact on the function of arbuscules. Since the symbiotic nutrient exchange largely occurs before the onset of stage IVt, this could be expected. On the other hand, an MtPt4-promoter controlled overexpression of MtDefMd1 led to higher MtRam1 transcription, indicating that an enhanced level of MtDefMd transcripts can lead to differential expression of a key transcription factor that invokes a faster development of the arbuscule interface (MtRam1) [16].
Interestingly, nodule-specific defensins were found in Alnus glutinosa root nodules [69]. AG5, one of these, caused physiological changes of Frankia vesicles and increased their permeability [69], whereas four additional defensins displayed partly overlapping functions [70]. Apart from classical non-symbiotic and symbiotic defensins, more than 700 defensin-like peptides containing N-terminal signal peptides and conserved cysteine residues were identified in legume plants [26]. These nodule-specific cysteine rich (NCR) peptides either contain 4 or 6 cysteines, and some of them display remarkable similarities with AM-related defensins. NCR-peptides are targeted to symbiosomes via the secretory pathway [35, 71]. Inside the lumen of nitrogen-fixing bacteroids, multiple targets of NCR-peptides were observed, e.g. ribosomal proteins, cell cyle regulators, ATP synthases, nitrogenase, and components of the TCA cycle [72, 73]. In concert, the effects of hundreds of cell-specific NCR-peptides lead to terminal bacteroid differentiation in the indeterminate-type legume root nodules [74].
Vacuolar V-SNARE receptors are recruited in early steps of bacteroid senescence [75, 76]. Cargo vesicles originally addressed to the vacuole are now delivered to degrading symbiosome membranes, transforming the enclosed perisymbiotic space to an acidic lytic compartment [77, 78]. Fragmented vacuoles are adaptations of mature arbuscule-containing cells [79], so these processes are of interest for AM as well. In addition, it was reported that similar to the peribacteroid space, the periarbuscular space is acidic [80]. This environment might be decisive for a protonation of the aspartic acid residue in the characteristic C-terminal region of mycorrhiza-related defensins, and this might be important for their function.
The massive formation of fluorophore-positive vacuoles at a specific point of the arbuscule life-cycle (Fig 8A, p and t) probably reflects the recycling of PAM-derived fatty acids and proteins by the host. Presence of large amounts of MtDefMd1-mGFP6 was not only confined to a rather short, but also to a very late moment in the arbuscule life-cycle (stage IVt.; Fig 8l, p and t,), a time point matched by the accumulation of oil droplets [15]. Since AM fungi are oleogenic, the discovery that lipid dropleds coincide with collapsed arbuscular branches [15] opens new perspectives for MtDefMd function.
Studies on MtRam1 and MtRam2 mutants demonstrated that in addition to sugars, fatty acids from the host are supplied to intraradical fungal structures [81, 82], thus compensating the lack of fungal multidomain fatty acid synthases [15, 83]. This direct supply of monoacylglycerol is probably an important source for the synthesis of triaglycerol stored in lipid bodies of the intraradical mycelium [84, 15]. The transfer of fatty acids probably occurs during the active, nutrient-exchanging phase of the arbuscule. At a later time point, arbuscules are regularly degraded, and this process generates another source of lipids. Although it is currently unknown, what mechanisms for the attachment of defensins to fungal membranes are used [30], it is intriguing that the Raphanus sativus AFP2 and the Dhahlia merckii AMP1 defensins are known to bind sphingolipids [31, 85, 30]. Although MtDefMd defensins exhibit some differences to non-symbiotic defensins, they are likely to have direct antifungal activity like other cationic, AM-unrelated defensins. In particular, AM-dependent defensins including their γ-core motifs share significant homologies with the recently published bi-domain MtDef5A/B, which exhibits potent antifungal activity in vitro [65]. Interestingly, MtDef5A/B has been shown to bind several phospholipids in vitro, including phosphatidylinositol monophosphates (PIP) and PIP2 [65]. It is thus likely that MtDefMd1 and MtDefMd2 also bind to phospholipids and this binding might be important for their biological function during later stage of the AM symbiosis. Taking the peak of MtDefMd1-mGFP6 synthesis in a short and late period of the arbuscule life-cycle into consideration, it is tempting to hypothesize that MtDefMd defensins bind specific lipid compounds during the massive digestion of degrading arbuscules, containing a significant amount of membrane lipids. During this process, these lipids might act as carriers for an endocytic incorporation of MtDefMd proteins, thereby limiting excessive fungal access to host lipids due to a potential toxicity of the defensins at high concentrations. This toxicity can e.g. be mediated via Ca2+-influx, K+-efflux, and a loss of membrane potential [86, 87, 88]. Since phytopathogenic fungi are supplied with host fatty acids as well, and reduced fatty acid synthesis impairs infections [82], mechanisms hindering the loss of plant fatty acids are an important mode of control.
The quick and strong accumulation of MtDefMd1 marks the initiation of a turnover in arbuscule-containing cells, ultimately leading to the post-symbiotic formation of cortical cells ready to be re-colonized. This transition requires not only transcriptional changes and cellular restructuring, but also antimicrobial treatments. Specifically, fungal structures collapse during the arbuscule degeneration initiated by MtMyb1 [17], while the now abandoned symbiotic market place is enriched with proteins and lipids of symbiotic membranes [14]. It might thus be beneficial for the plant to recycle such molecules by redirecting them to the vacuole that grows due to the fusion of initially fragmented areas. Absence of MtDefMd1 and MtDefMd2 transcription in pt4-2 and ram1-1 mutants has thus to be expected, since in defect or prematurely degraded arbuscules, no expanded symbiotic interface exists, where lipids need to be reconquered.
Due to the multiplity of their modes of action, functional assessments of defensin-like proteins are challenging. We here show that MtDefMd1, a member of the newly described MtDefMd family of mycorrhiza-activated defensins, is not only specifically transcribed during AM interactions, but is present during the restructuring processes of arbuscule-containing cells. Its presence at this defined, late stage of the arbuscule life-cycle might be a hallmark of the regular transition from symbiotic to post-symbiotic cells. Since the timing of defensin presence described here coincides with the reported accumulation of AM-fungal lipids [15], an association of AM-related defensins with these molecules is a starting point for furthers studies of MtDefMd function in the AM symbiosis.
Supporting information
Confocal micrographs of razor blade hand-cuttings of transgenic M. truncatula roots. The roots express an MtDefMd1-mGFP6 fusion under the control of the native promoter (A; d and g), an ER-CFP fusion under the control of the 2x35S-promoter (A; c and f), and a fusion of the signal peptide of MtBcp1 with mCherry under the control of the native promoter (A; b). Additionally, a tonoplast membrane directed GFP fusion under the control of a 2x35S-promoter (B, b) is shown. Differential interference contrast (DIC) micrographs are shown for each root section (A; a and e; B, a). Roots were mycorrhized with R. irregularis for six weeks.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Manfred K. Schenk (Institute of Plant Nutrition, Leibniz Universität Hannover, Hannover, Germany) for helpful advice on the setup of plant nutrition solutions and Natascha Köppens (Institut of Plant Genetics, Leibniz Universität Hannover, Hannover, Germany) for excellent technical assistance. Medicago truncatula pt4-2 and ram1-1 seeds were kindly provided by Maria Harrison (Boyce Thompson Institute, Ithaca, NY, USA) and Giles Oldroyd (John Innes Centre, Norwich, UK), respectively. We are grateful to Bettina Hause (IPB, Halle, Germany) for supplying AM fungal inoculum, to Miriam Laxa (Institut für Botanik, Leibniz Universität Hannover, Hannover, Germany) for the cell organelle localization vectors pBIN:tp-gk and pBIN:er-ck, to Franziska Krajinski-Barth (Institut für Biologie, Universität Leipzig, Leipzig, Germany) for the overexpression vectors 315p9RFP-Pt4-Expr and 917p9RFP-ubi3-Expr, to Erik Limpens (Department of Plant Sciences, Wageningen University, Wageningen, The Netherlands) for the RNAi vector pK7GWIWG2(II)-Q10:DsRED, and to Maria Harrison (Boyce Thompson Institute, Ithaca, NY, USA) for the MtBcp1 localization vector pCMbB-TMEr.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors wish to thank the Deutsche Forschungsgemeinschaft (http://www.dfg.de) for financial support in frame of GRK1798 "Signaling at the Plant-Soil Interface". The publication of this article was funded by the Open Access Fund of the Leibniz Universität Hannover. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Charpentier M, Bredemeier R, Wanner G, Takeda N, Schleiff E, and Parniske M. Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 2008; 20(12): 3467–3479 10.1105/tpc.108.063255] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schüssler A, Schwarzott D, and Walker C. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 2001; 105(12):1413–1421 [Google Scholar]
- 3.Bago B. Carbon export from arbuscular mycorrhizal roots involves the translocation of carbohydrate as well as lipid. Plant Physiol 2003; 131(3): 1496–1507 10.1104/pp.102.007765] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wewer V, Brands M, and Dörmann P. Fatty acid synthesis and lipid metabolism in the obligate biotrophic fungus Rhizophagus irregularis during mycorrhization of Lotus japonicus. Plant Journal 2014; 79(3): 398–412 10.1111/tpj.12566] ] [DOI] [PubMed] [Google Scholar]
- 5.Smith SE and Smith FA. Roles of arbuscular mycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Annu Rev Plant Biol 2011; 62(1): 227–250. 10.1146/annurev-arplant-042110-103846] ] [DOI] [PubMed] [Google Scholar]
- 6.Simon L, Bousquet J, Lévesque RC, and Lalonde M. Origin and diversification of endomycorrhizal fungi and coincidence with vascular land plants. Nature 1993; 363(6424): 67–69 10.1038/363067a0] [DOI] [Google Scholar]
- 7.Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, and Hedges SB. Molecular evidence for the early colonization of land by fungi and plants. Science 2001; 293(5532): 1129–1133 10.1126/science.1061457] ] [DOI] [PubMed] [Google Scholar]
- 8.Akiyama K, Matsuzaki Ki, and Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005; 435(7043): 824–827 10.1038/nature03608] [DOI] [PubMed] [Google Scholar]
- 9.Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, et al. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol 2006; 4(7): e226 10.1371/journal.pbio.0040226] ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison MJ. The arbuscular mycorrhizal symbiosis: An underground association. Trends Plant Sci 1997; 2(2): 54–60 10.1016/S1360-1385(97)82563-0] [DOI] [Google Scholar]
- 11.Strack D, Fester T, Hause B, Schliemann W, and Walter MH. Arbuscular mycorrhiza: Biological, chemical, and molecular aspects. J Chem Ecol 2003; 29(9): 1955–1979 ] [DOI] [PubMed] [Google Scholar]
- 12.Genre A, Chabaud M, Timmers T, Bonfante P, and Barker DG. Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 2005; 17(12): 3489–3499 10.1105/tpc.105.035410] ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox G and Tinker PB. Translocation and transfer of nutrients in vesicular-arbuscular mycorrhizas. I.The arbuscule and phosphorous transfer: A quantitative ultrastructural study. New Phytol 1976; 77(2): 371–378 10.1111/j.1469-8137.1976.tb01526.x] [DOI] [Google Scholar]
- 14.Kobae Y and Hata S. Dynamics of periarbuscular membranes visualized with a fluorescent phosphate transporter in arbuscular mycorrhizal roots of rice. Plant Cell Phys 2010; 51(3): 341–353 10.1093/pcp/pcq013] ] [DOI] [PubMed] [Google Scholar]
- 15.Kobae Y, Gutjahr C, Paszkowski U, Kojima T, Fujiwara T, and Hata S. Lipid droplets of arbuscular mycorrhizal fungi emerge in concert with arbuscule collapse. Plant Cell Phys 2014; 55(11): 1945–1953 10.1093/pcp/pcu123] ] [DOI] [PubMed] [Google Scholar]
- 16.Park HJ, Floss DS, Levesque-Tremblay V, Bravo A, and Harrison MJ. Hyphal branching during arbuscule development requires reduced arbuscular mycorrhiza 1. Plant Physiol 2015; 169(4):2774–2788 10.1104/pp.15.01155] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Floss DS, Gomez SK, Park HJ, MacLean AM, Müller LM, Bhattarai KK, et al. A transcriptional program for arbuscule degeneration during AM symbiosis is regulated by MYB1. Current Biol 2017; 27(8): 1206–1212 10.1016/j.cub.2017.03.003] [DOI] [PubMed] [Google Scholar]
- 18.Hogekamp C and Küster H. A roadmap of cell-type specific gene expression during sequential stages of the arbuscular mycorrhiza symbiosis. BMC Gen 2013; 14(1): 306 10.1186/1471-2164-14-306] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hohnjec N, Vieweg MF, Pühler A, Becker A, and Küster H. Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol 2005; 137(4): 1283–1301 10.1104/pp.104.056572] ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Provorov NA, Borisov AY, and Tikhonovich IA. Developmental genetics and evolution of symbiotic structures in nitrogen-fixing nodules and arbuscular mycorrhiza. J of Theoretical Biol 2002; 214(2): 215–232 10.1006/jtbi.2001.2453 [DOI] [PubMed] [Google Scholar]
- 21.Ivanov S, Fedorova EE, Limpens E, De Mita S, Genre A, Bonfante P, et al. Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc Natl Acad Sci U S A 2012; 109(21): 8316–8321 10.1073/pnas.1200407109] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Pumplin N, Ivanov S, and Harrison MJ. EXO70I is required for development of a sub-domain of the periarbuscular membrane during arbuscular mycorrhizal symbiosis. Current Biol 2015; 25(16): 2189–2195 10.1016/j.cub.2015.06.075] [DOI] [PubMed] [Google Scholar]
- 23.Harrison MJ and Ivanov S. Exocytosis for endosymbiosis: membrane trafficking pathways for development of symbiotic membrane compartments. Current Op Plant Biol 2017; 38:101–108 10.1016/j.pbi.2017.04.019] [DOI] [PubMed] [Google Scholar]
- 24.Huisman R, Hontelez J, Mysore KS, Wen J, Bisseling T, and Limpens E. A symbiosis-dedicated syntaxin of plants 13II isoform controls the formation of a stable host-microbe interface in symbiosis. New Phytol 2016; 211(4): 1338–1351 10.1111/nph.13973 [DOI] [PubMed] [Google Scholar]
- 25.Chen JS, Reddy V., Chen JH, Shlykov MA, Zheng WH, Cho J, et al. Phylogenetic characterization of transport protein superfamilies: superiority of SuperfamilyTree programs over those based on multiple alignments. J. Mol Microbiol. Biotechnol (2011). 21(3–4): 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maróti G, Downie JA, and Kondorosi É. Plant cysteine-rich peptides that inhibit pathogen growth and control rhizobial differentiation in legume nodules. Current Op Plant Biol 2015; 26: 57–63 10.1016/j.pbi.2015.05.031 [DOI] [PubMed] [Google Scholar]
- 27.Thomma BPHJ, Cammue BPA, and Thevissen K. Plant defensins. Planta 2002; 216(2): 193–202 10.1007/s00425-002-0902-6 [DOI] [PubMed] [Google Scholar]
- 28.García-Olmedo F, Molina A, Alamillo JM, and Rodríguez-Palenzuéla P. Plant defense peptides. Biopolymers 1998; 47(6): 479– [DOI] [PubMed] [Google Scholar]
- 29.Van der Weerden NL and Anderson MA. Plant defensins: common fold, multiple functions. Fungal Biol Rev 2013; 26(4):121–131 10.1128/AAC.00365-13 [DOI] [Google Scholar]
- 30.Sagaram US, El-Mounadi K, Buchko GW, Berg HR, Jagdeep Kaur J, Pandurangi RS, et al. Structural and functional studies of a phosphatidic acid-binding antifungal plant defensin MtDef4: Identification of an RGFRRR motif governing fungal cell entry. PLoS ONE 2013; 8(12): e82485 10.1371/journal.pone.0082485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thevissen K, Osborn RW, Acland DP, and Broekaert WF. Specific binding sites for an antifungal plant defensin from Dahlia (Dahlia merckii) on fungal cells are required for antifungal activity. Mol Plant Microbe Interact 2000; 13(1): 54–61 10.1094/MPMI.2000.13.1.54 [DOI] [PubMed] [Google Scholar]
- 32.Kagan BL, Selsted ME, Ganz T, and Lehrer RI. Antimicrobial defensin peptides form voltage-dependent ion-permeable channels in planar lipid bilayer membranes. Proc Natl Acad Sci U S A 1990; 87(1): 210–214 10.1073/pnas.87.1.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanks JN, Snyder AK, Graham MA, Shah RK, Blaylock LA, Harrison MJ, et al. Defensin gene family in Medicago truncatula: Structure, expression and induction by signal molecules. Plant Mol Biol 2005; 58(3): 385–399 10.1007/s11103-005-5567-7 [DOI] [PubMed] [Google Scholar]
- 34.Alunni B and Gourion B. Terminal bacteroid differentiation in the legume−Rhizobium symbiosis: nodule-specific cysteine-rich peptides and beyond. New Phytol 2016; 211(2):411–417 10.1111/nph.14025 [DOI] [PubMed] [Google Scholar]
- 35.van de Velde W, Zehirov G, Szatmari A Debreczeny M, Ishihara H, Kevei Z, et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 2010; 327(5969): 1122–1126 10.1126/science.1184057 [DOI] [PubMed] [Google Scholar]
- 36.Gomez SK, Javot H, Deewatthanawong P, Ivone Torres-Jerez I, Tang Y, Blancaflor EB,et al. Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol 2009; 9(1): 10 10.1186/1471-2229-9-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hogekamp C, Arndt D, Pereira PA, Becker JD, Hohnjec N, and Küster H. Laser microdissection unravels cell-type-specific transcription in arbuscular mycorrhizal roots, including CAAT-Box transcription factor gene expression correlating with fungal contact and spread. Plant Physiol 2011; 157(4): 2023–2043 10.1104/pp.111.186635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaude N, Bortfeld S, Duensing N, Lohse M, and Krajinski F. Arbuscule-containing and non-colonized cortical cells of mycorrhizal roots undergo extensive and specific reprogramming during arbuscular mycorrhizal development. Plant Journal 2012; 69(3): 510–528 10.1111/j.1365-313X.2011.04810.x [DOI] [PubMed] [Google Scholar]
- 39.Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, et al. Template-based protein structure modeling using the RaptorX web server. Nat Protoc 2012; 7(8): 1511–1522 10.1038/nprot.2012.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vriens K, Cools TL, Harvey PJ, Craik DJ, Spincemaille P, Cassiman D, et al. Synergistic activity of the plant defensin HsAFP1 and caspofungin against Candida albicans biofilms and planktonic cultures. PLoS ONE 2015; 10(8): e0132701 10.1371/journal.pone.0132701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fant F, Vranken W, Broekaert W, and Borremans F. Determination of the three-dimensional solution structure of Raphanus sativus Antifungal Protein 1 by 1H NMR11. J of Mol Biol 1998, 279 (1): 257–270 10.1006/jmbi.1998.1767. [DOI] [PubMed] [Google Scholar]
- 42.Razzera G, Gadermaier G, de Paula V, Almeida MS, Egger M, Jahn-Schmid B, et al. Mapping the interactions between a major pollen allergen and human IgE antibodies. Structure 2010; 18(8): 1011–1021 10.1016/j.str.2010.05.012 [DOI] [PubMed] [Google Scholar]
- 43.Krishnakumar V, Kim M, Rosen BD, Karamycheva S, Bidwell SL, Tang H, et al. MTGD: The Medicago truncatula genome database. Plant Cell Physiol 2015; 56(1):e1 10.1093/pcp/pcu179 [DOI] [PubMed] [Google Scholar]
- 44.Tang H, Krishnakumar V, Bidwell S, Rosen B, Chan A, Zhou S, et al. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Gen 2014; 15: 312 10.1186/1471-2164-15-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petersen T, Brunak S, von Heijne G, and Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods 2011; 8(10):785–786 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 46.Chou KC and Shen HB. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010; 5(6): e11335 10.1371/journal.pone.0011335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Briesemeister S, Blum T, Brady S, Lam Y, Kohlbacher O, and Shatkay H. SherLoc2: A high-accuracy hybrid method for predicting subcellular localization of proteins. J Proteome Res 2009; 8(11): 5363–5366 10.1021/pr900665y [DOI] [PubMed] [Google Scholar]
- 48.Grant SG, Jessee J, Bloom FR, and Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A 1990; 87(12): 4645–4649 10.1073/pnas.87.12.4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quandt HJ. Transgenic root nodules of Vicia hirsuta: A fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 1993; 6(6): 699 10.1094/MPMI-6-699 [DOI] [Google Scholar]
- 50.Pridmore RD. New and versatile cloning vectors with kanamycin-resistance marker. Gene 1987; 56(2–3): 309–312 10.1016/0378-1119(87)90149-1 [DOI] [PubMed] [Google Scholar]
- 51.Küster H, Quandt HJ, Broer I, Perlick AM, and Pühler A. The promoter of the Vicia faba L. VfENOD-GRP3 gene encoding a glycine-rich early nodulin mediates a predominant gene expression in the interzone II-III region of transgenic Vicia hirsuta root nodules. Plant Mol Biol 1995; 29(4): 759–772 10.1007/BF00041166 [DOI] [PubMed] [Google Scholar]
- 52.Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, et al. RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 2004; 55(399): 983–992 [DOI] [PubMed] [Google Scholar]
- 53.Limpens E, Mirabella R, Fedorova E, Franken C, Franssen H, Bisseling T, et al. Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc Natl Acad Sci U S A 2005; 102(29): 10375–10380 10.1073/pnas.0504284102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Devers EA, Teply J, Reinert A, Gaude N, and Krajinski F. An endogenous artificial microRNA system for unraveling the function of root endosymbioses related genes in Medicago truncatula. BMC Plant Biol 2013; 13: 82 10.1186/1471-2229-13-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harrison MJ, Dewbre GR, and Liu J. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 2002; 14(10): 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baier M, Hohnjec N, Lenz F, Fehlberg V, Vieweg MF, Hause B, et al. The signal peptide of the Medicago truncatula modular nodulin MtNOD25 operates as an address label for the specific targeting of proteins to nitrogen-fixing symbiosomes. Mol Plant Microbe Interact 2009; 22(1): 63–72 10.1094/MPMI-22-1-0063 [DOI] [PubMed] [Google Scholar]
- 57.Nelson BK, Cai X, and Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant Journal 2007; 51(6): 1126–1136 10.1111/j.1365-313X.2007.03212.x [DOI] [PubMed] [Google Scholar]
- 58.Ivanov S and Harrison MJ. A set of fluorescent protein-based markers expressed from constitutive and arbuscular mycorrhiza-inducible promoters to label organelles, membranes and cytoskeletal elements in Medicago truncatula. Plant Journal 2014; 80(6): 1151–1163 10.1111/tpj.12706 [DOI] [PubMed] [Google Scholar]
- 59.Arnon DI and Hoagland DR. Crop production in artificial culture solutions and in soils with special reference to factors influencing yields and absorption of inorganic nutrients. Soil Sci. 1940; 50: 463–483. [Google Scholar]
- 60.Vieweg MF, Frühling M, Quandt H-J, Heim U, Bäumlein H, Pühler A, et al. The promoter of the Vicia faba L. leghemoglobin gene VfLb29 is specifically activated in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots from different legume and nonlegume plants. Mol Plant Microbe Interact 2004; 17(1): 62–69 10.1094/MPMI.2004.17.1.62 [DOI] [PubMed] [Google Scholar]
- 61.Javot H, Penmetsa RV, Breuillin F, Bhattarai KK, Noar RD, Gomez SK, et al. Medicago truncatula mtpt4 mutants reveal a role for nitrogen in the regulation of arbuscule degeneration in arbuscular mycorrhizal symbiosis. Plant Journal 2011; 68(6): 954–965 10.1111/j.1365-313X.2011.04746.x [DOI] [PubMed] [Google Scholar]
- 62.Brundrett M, Bougher N, Dell B,Grove T, and Malajczuk N. Working with mycorrhizas in forestry and agriculture ACIAR monograph 1996; ACIAR: Canberra: Publication Code: MN032ISBN: 1 86320 181 5 [Google Scholar]
- 63.Benedito VA, Torres-Jerez I, Murray JD, et al. A gene expression atlas of the model legume Medicago truncatula. Plant Journal 2008; 55(3): 504–513 10.1111/j.1365-313X.2008.03519.x [DOI] [PubMed] [Google Scholar]
- 64.Pumplin N, Zhang X, Noar RD, and Harrison MJ. Polar localization of a symbiosis-specific phosphate transporter is mediated by a transient reorientation of secretion. Proc Natl Acad Sci U S A 2012; 109(11): E665–672 10.1073/pnas.1110215109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Islam KT, Velivelli SLS, Berg RH, Oakley B, and Shah DM. A novel bi-domain plant defensin MtDef5 with potent broad-spectrum antifungal activity binds to multiple phospholipids and forms oligomers. Sci Rep 2017; 7(1): 135 10.1038/s41598-017-16508-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Javot H, Penmetsa RV, Terzaghi N, Cook DR, and Harrison MJ. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci U S A 2007; 104(5): 1720–1725 10.1073/pnas.0608136104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonneau L, Huguet S, Wipf D, Pauly N, and Truong HN. Combined phosphate and nitrogen limitation generates a nutrient stress transcriptome favorable for arbuscular mycorrhizal symbiosis in Medicago truncatula. New Phytol 2013; 199(1): 188–202 10.1111/nph.12234 [DOI] [PubMed] [Google Scholar]
- 68.Gobbato E, Marsh JF, Vernié T, Wang E, Maillet F, Kim J, et al. A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Current Biol 2012; 22(23): 2236–2241 10.1016/j.cub.2012.09.044 [DOI] [PubMed] [Google Scholar]
- 69.Carro L, Pujic P, Alloisio N, Fournier P, Boubakri H, Hay AE, et al. Alnus peptides modify membrane porosity and induce the release of nitrogen-rich metabolites from nitrogen-fixing Frankia. ISME J 2015; 9(8): 1723–1733 10.1038/ismej.2014.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carro L, Pujic P, Alloisio N, Fournier P, Boubakri H, Poly F, et al. Physiological effects of major up-regulated Alnus glutinosa peptides on Frankia sp. ACN14a. Microbiology 2016; 162(7): 1173–1184 10.1099/mic.0.000291 [DOI] [PubMed] [Google Scholar]
- 71.Mergaert P. A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 2003; 132(1): 161–173 10.1104/pp.102.018192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Penterman J, Abo RP, De Nisco NJ, Arnold MFF, Longhi R, Zanda M, et al. Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. Proc Natl Acad Sci U S A 2014; 111(9): 3561–3566 10.1073/pnas.1400450111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Farkas A, Maróti G, Durgő H, Györgypál Z, Lima RM, Medzihradszky KF, et al. Medicago truncatula symbiotic peptide NCR247 contributes to bacteroid differentiation through multiple mechanisms. Proc Natl Acad Sci U S A 2014; 111(14): 5183–5188 10.1073/pnas.1404169111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maróti G. and Kondorosi É. Nitrogen-fixing Rhizobium-legume symbiosis: Are polyploidy and host peptide-governed symbiont differentiation general principles of endosymbiosis? Front Microbiol. 2014; 5(359): 636 10.3389/fmicb.2014.00326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Limpens E, Ivanov S, van Esse W, Voets G, Fedorova E, and Bisseling T. Medicago N2-fixing symbiosomes acquire the endocytic identity marker Rab7 but delay the acquisition of vacuolar identity. Plant Cell 2009; 21(9): 2811–2828 10.1105/tpc.108.064410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pierre O, Engler G, Hopkins J, Brau F, Boncompagni E, and Hérouart D. Peribacteroid space acidification: A marker of mature bacteroid functioning in Medicago truncatula nodules. Plant Cell Environ 2013; 36(11): 2059–2070 10.1111/pce.12116 [DOI] [PubMed] [Google Scholar]
- 77.Pladys D and Vance CP. Proteolysis during development and senescence of effective and plant gene-controlled ineffective alfalfa nodules. Plant Physiol 1993; 103(2): 379–384 10.1104/pp.103.2.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van de Velde W, Guerra JCP, De Keyser A, De Rycke R, Rombauts S, Maunoury N, et al. Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula. Plant Physiol 2006; 141(2): 711–720 10.1104/pp.106.078691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hause B and Fester T. Molecular and cell biology of arbuscular mycorrhizal symbiosis. Planta 2005; 221(2): 184–196 10.1007/s00425-004-1436-x [DOI] [PubMed] [Google Scholar]
- 80.Krajinski F, Courty PE, Sieh D, Franken P, Zhang H, Bucher M, et al. The H+-ATPase HA1 of Medicago truncatula is essential for phosphate transport and plant growth during arbuscular mycorrhizal symbiosis. Plant Cell 2014; 26(4): 1808–1817 10.1105/tpc.113.120436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luginbuehl LH, Menard GN, Kurup S, Van Erp H, Radhakrishnan GV, Breakspear A, et al. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 2017; 356(6343): 1175–1178 10.1126/science.aan0081 [DOI] [PubMed] [Google Scholar]
- 82.Jiang Y, Wang W, Xie Q, Liu N, Liu L, Wang D, et al. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 2017; 356(6343): 1172–1175 10.1126/science.aam9970 [DOI] [PubMed] [Google Scholar]
- 83.Tisserant E, Malbreil M, Kuo A, Annegret Kohler A, Symeonidi A, Balestrini R, et al. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci U S A 2013; 110(50): 20117–20122 10.1073/pnas.1313452110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bago B. Translocation and utilization of fungal storage lipid in the arbuscular mycorrhizal symbiosis. Plant Physiol 2002; 128(1): 108–124 10.1104/pp.128.1.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thevissen K, De Mello Tavares P, Xu D, Blankenship J, Vandenbosch D, Idkowiak-Baldys J, et al. The plant defensin RsAFP2 induces cell wall stress, septin mislocalization and accumulation of ceramides in Candida albicans. Mol Microbiol 2012; 84(1): 166–180 10.1111/j.1365-2958.2012.08017.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hayes BME, Bleackley MR, Wiltshire JL, Anderson MA, Traven A, and van der Weerden NL. Identification and mechanism of action of the plant defensin NaD1 as a new member of the antifungal drug arsenal against Candida albicans. Antimicrob Agents Chemother 2013; 57(8): 3667–3675 10.1128/AAC.00365-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lobo DS, Pereira IB, Fragel-Madeira L, Medeiros LN, Cabral LM, Faria J, et al. Antifungal Pisum sativum defensin 1 interacts with Neurospora crassa cyclin F related to the cell cycle. Biochemistry 2007; 46(4): 987–996 10.1021/bi061441j [DOI] [PubMed] [Google Scholar]
- 88.van der Weerden NL, Lay FT, and Anderson MA. The plant defensin, NaD1, enters the cytoplasm of Fusarium oxysporum hyphae. J Biol Chem 2008; 283(21): 14445–14452 10.1074/jbc.M709867200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confocal micrographs of razor blade hand-cuttings of transgenic M. truncatula roots. The roots express an MtDefMd1-mGFP6 fusion under the control of the native promoter (A; d and g), an ER-CFP fusion under the control of the 2x35S-promoter (A; c and f), and a fusion of the signal peptide of MtBcp1 with mCherry under the control of the native promoter (A; b). Additionally, a tonoplast membrane directed GFP fusion under the control of a 2x35S-promoter (B, b) is shown. Differential interference contrast (DIC) micrographs are shown for each root section (A; a and e; B, a). Roots were mycorrhized with R. irregularis for six weeks.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.