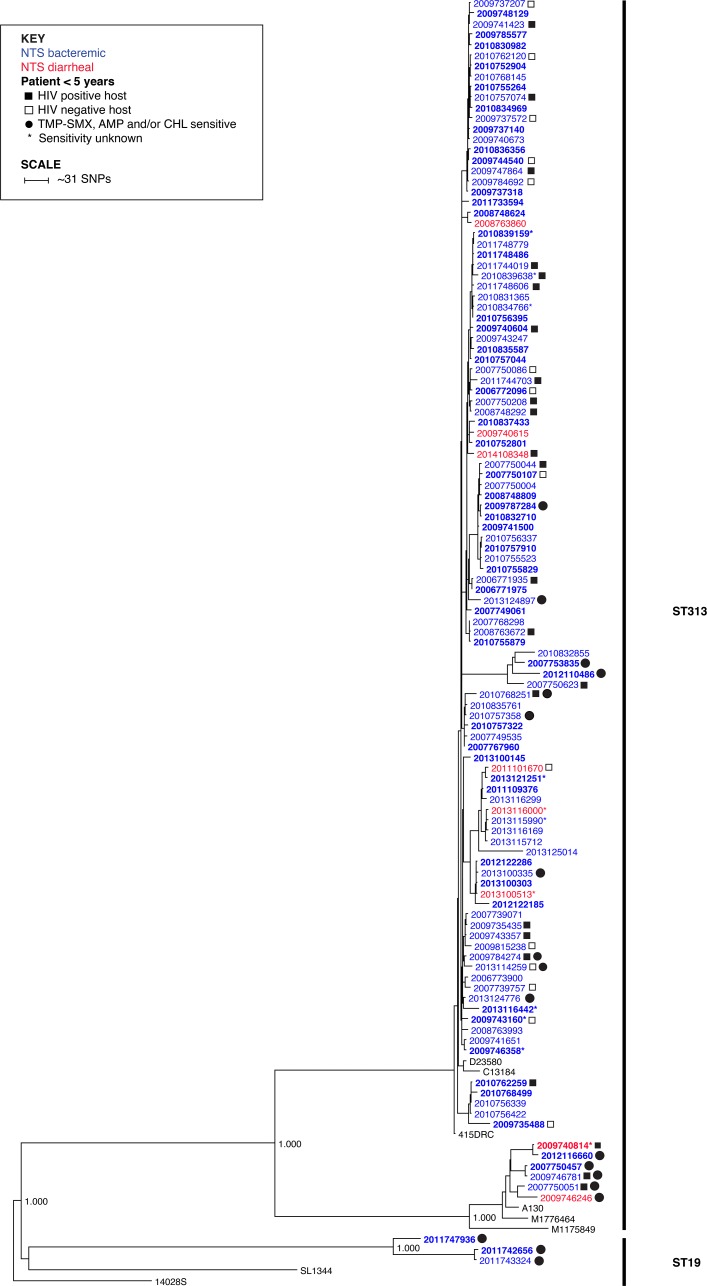
Fig 2. Maximum likelihood phylogeny of 115 S. Typhimurium strains (3 strains are from the same patient at separate visits within one month and are not included in the epidemiologic analysis) based on k-mers.
Each strain is represented by collection year for clarity (strain numbers can be found in the S1 Table). NTS bacteremic and diarrheal clinical strains are colored blue and red, respectively. Reference strains appear in black text. Bold text indicates strains collected from patients under 5 years of age. Filled and open squares indicate HIV positive and negative hosts, respectively, when known. Clinical strains susceptible to TMP-SMX, ampicillin (AMP) and/or chloramphenicol (CHL) are indicated with filled circles. Strains with unknown susceptibilities are indicated with an asterisk (*). Basal branches with 100% local support are labeled. The tree was rooted with laboratory strain 14028S.