Figure 2.
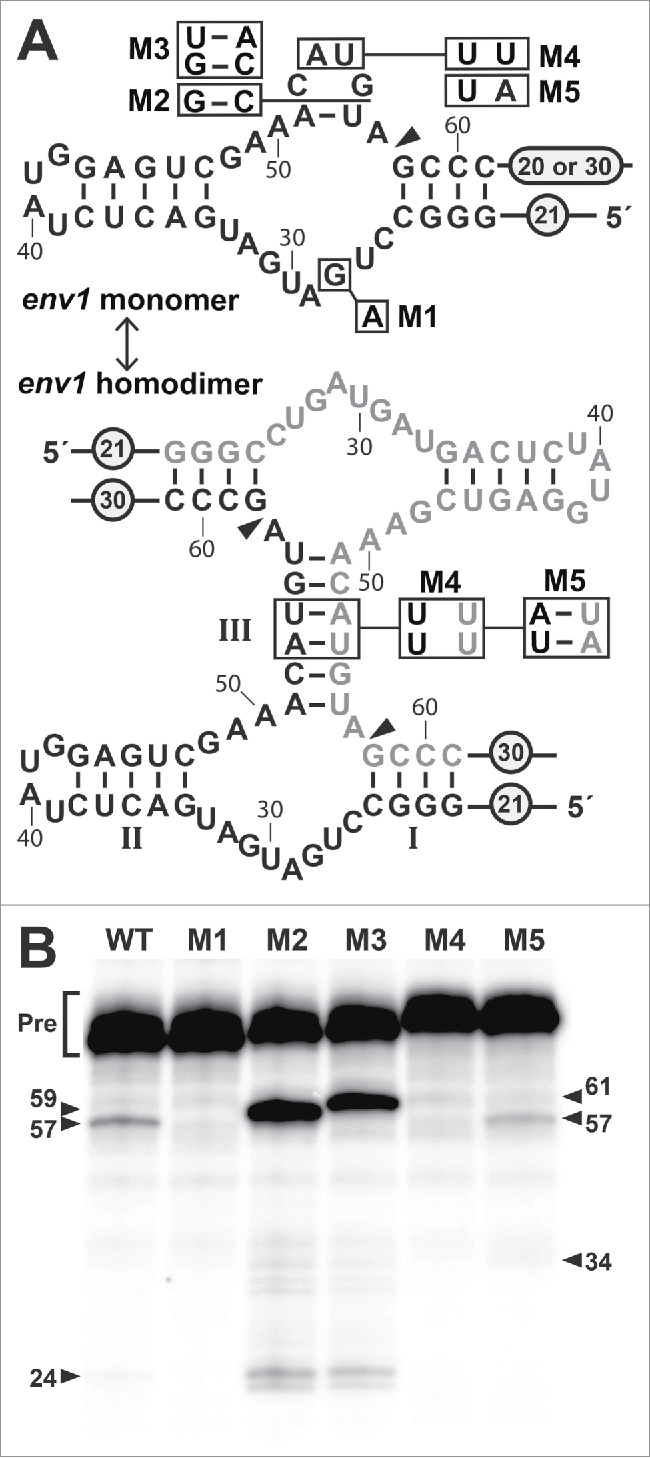
Sequence, predicted secondary structures, and functional characteristics of env1, a representative SSIII hammerhead. (A) Sequence and secondary structure prediction for a single WT env1 RNA (top) and 2 env1 RNAs forming a dimer (bottom). Constructs carrying mutations at specific sites (boxed) are designated M1 through M5. Note that M2 and M3 are insertions at the location indicated by the line. Encircled numbers indicate the length of added native nucleotides surrounding the conserved ribozyme core. 20 nucleotides were present on the 3′ terminus of the WT, M1, M2 and M3 constructs, whereas 30 nucleotides were present on constructs M4 and M5. Arrowhead designates the site of cleavage, and nucleotide numbering is relative to the in vitro transcription start site. (B) Co-transcriptional cleavage of WT env1 and its mutants was monitored by denaturing polyacrylamide gel electrophoresis (PAGE) of internally 32P-labeled RNA (α-32P-GTP) (see Materials and Methods for details). Full-length precursor (Pre) RNAs in nucleotides are 82 (WT and M1), 84 (M2), 86 (M3) and 92 (M4 and M5). Ribozyme cleavage product bands in nucleotides are 57 and 24 (WT and M1), 59 and 24 (M2), 61 and 24 (M3), and 57 and 35 (M4 and M5) as annotated with numbered arrowheads.
