Figure 3.
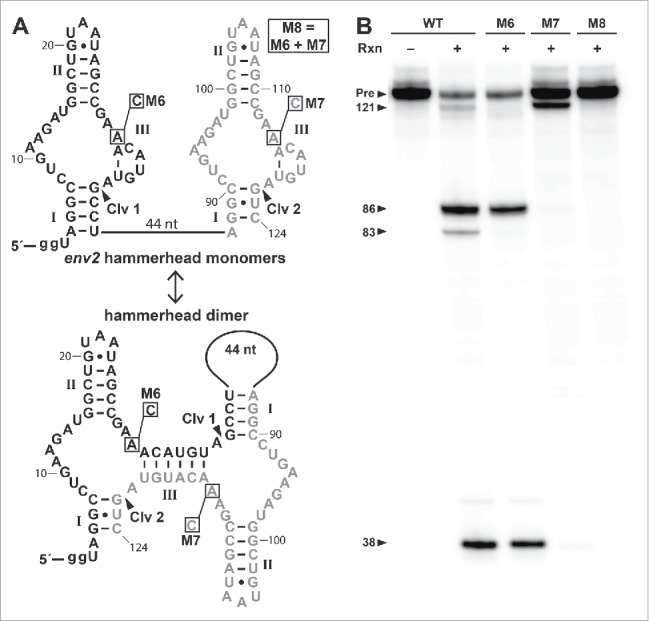
A tandem hammerhead cleaves as an obligate dimer. (A) Sequence and secondary structures of the WT tandem hammerhead construct env2 depicted as independently folded (top) and dimer (bottom) structures. Hammerhead depiction is rotated 90° from previous figures. Individual ribozyme sequences are shown in black (first ribozyme) and gray (second ribozyme). Mutations to active-site nucleotides in constructs M6, M7 and M8 are boxed. Two guanine nucleotides on the 5′ end were added for efficient transcription (see Materials and Methods). Other annotations are as described for Fig. 2A. (B) Ribozyme cleavage reactions (Rxn) were conducted (+) at 37°C for 2 h using 50 mM Tris-HCl (pH 8.0 at 23°C), 0.5 mM EDTA, 10 mM MgCl2 and internally labeled RNA. The no reaction lane (−) yields only the full length RNA precursor (Pre). The expected 3 nt RNA cleavage fragment has been run off the gel.
