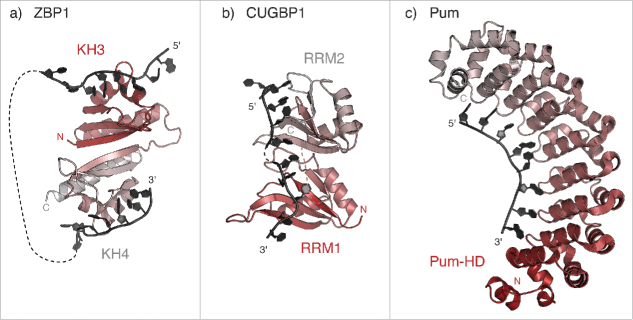
Figure 2.
Recognition of linear localization signals. (a) Model of RNA binding by KH3 and KH4 domains of the human ZBP1 homolog IMP1 (PDB 3KRM37), showing the pseudo-dimer arrangement that positions the RNA-binding surfaces on opposing directions of the structure. The RNA molecules bound to KH3 and KH4 are derived from the structure of Gallus ZBP1 (PDB 2N8L and 2N8M, respectively128), superposed using the cealign command in Pymol v1.7 (www.pymol.org). (b) Model of RNA binding by RRM1 and RRM2 domains of human CUGBP1 (PDB 3NNH and 3NMR, respectively). The relative orientation of the two RRM domains is modeled as described in Teplova et al., 200938. (c) Crystal structure of Drosophila Pum in complex with the Pum Recognition Element (PRE) from hunckback (hb) mRNA (PDB 5KLA76).