Abstract
HIF-1α, one of the most extensively studied oncogenes, is activated by a variety of microenvironmental factors. The resulting biological effects are thought to depend on its transcriptional activity. The RNAse enzyme Dicer is frequently downregulated in human cancers, which has been functionally linked to enhanced metastatic properties; however, current knowledge of the upstream mechanisms regulating Dicer is limited. In the present study, we identified Dicer as a HIF-1α–interacting protein in multiple types of cancer cell lines and different human tumors. HIF-1α downregulated Dicer expression by facilitating its ubiquitination by the E3 ligase Parkin, thereby enhancing autophagy-mediated degradation of Dicer, which further suppressed the maturation of known tumor suppressors, such as the microRNA let-7 and microRNA-200b. Consequently, expression of HIF-1α facilitated epithelial-mesenchymal transition (EMT) and metastasis in tumor-bearing mice. Thus, this study uncovered a connection between oncogenic HIF-1α and the tumor-suppressive Dicer. This function of HIF-1α is transcription independent and occurs through previously unrecognized protein interaction–mediated ubiquitination and autophagic proteolysis.
Keywords: Cell Biology
Keywords: Autophagy, Molecular biology, Ubiquitin-proteosome system
Introduction
HIF-1α is one of the best-studied oncogenes and it transcriptionally regulates genes to facilitate cancer metastasis (1, 2). Although HIF-1α accumulation was originally identified under hypoxia, as implied by its name (3, 4), a large body of research indicates that HIF-1α is strongly activated by microenvironmental stimulations under well-oxygenated conditions, such as serum starvation, IFN-α, chemotherapeutic drug treatments, and the most-studied IGF and EGF stimulations of growth factors (1). These studies indicate that HIF-1α is a central molecular hub for a variety of microenvironmental stimulations (5). During the process of cancer progression, tumor cells may develop an increased metastatic ability. To regulate downstream genes, HIF-1α undergoes heterodimerization with HIF-1β and then recognizes the HIF-1α–response elements in the promoters of specific genes (1, 6–8). The transcriptional complex activates a subset of genes, thus enhancing the malignant progression of cancer (1).
MicroRNAs (miRNAs) are critical gene regulators that are initially transcribed by RNA polymerase II and sequentially processed into mature miRNAs. Nascent primary miRNAs are processed in the cell nucleus by the RNase enzyme Drosha to become pre-miRNAs with a hairpin structure, which are then exported to the cytoplasm for a final maturation step processed by Dicer, a highly conserved RNase III enzyme that is expressed in eukaryotes and is essential for miRNA biogenesis. Mature miRNAs are eventually loaded into the miRNA-induced silencing complex (miRISC) to recognize the 3′ UTRs of target mRNAs (9, 10). Global repression of miRNAs in human cancers has been observed in previous studies (11, 12) and has been suggested to result from defective biogenesis machinery. Dysregulation of Dicer is of great interest because, similarly to the suppression of miRNA, downregulation of Dicer has been widely observed in many types of human tumors and Dicer has been shown to function as a tumor suppressor (12–17). Suppression of Dicer expression was found to dramatically enhance cancer metastasis through miR-200 miRNA (15, 18). These previous studies have identified the tumor-suppressive function of Dicer, but the upstream mechanism leading to Dicer downregulation, especially on the regulation of protein degradation, remains largely unknown. Using genetic manipulations and biological stimulations, we uncover a mechanism through which, under either hypoxic or nonhypoxic conditions, HIF-1α posttranslationally facilitates the ubiquitination and autophagic proteolysis of Dicer through direct protein-protein interaction, regardless of its canonical transcriptional activity.
Results
HIF-1α directly interacts with Dicer in multiple types of human cancer cell lines and tumor tissues.
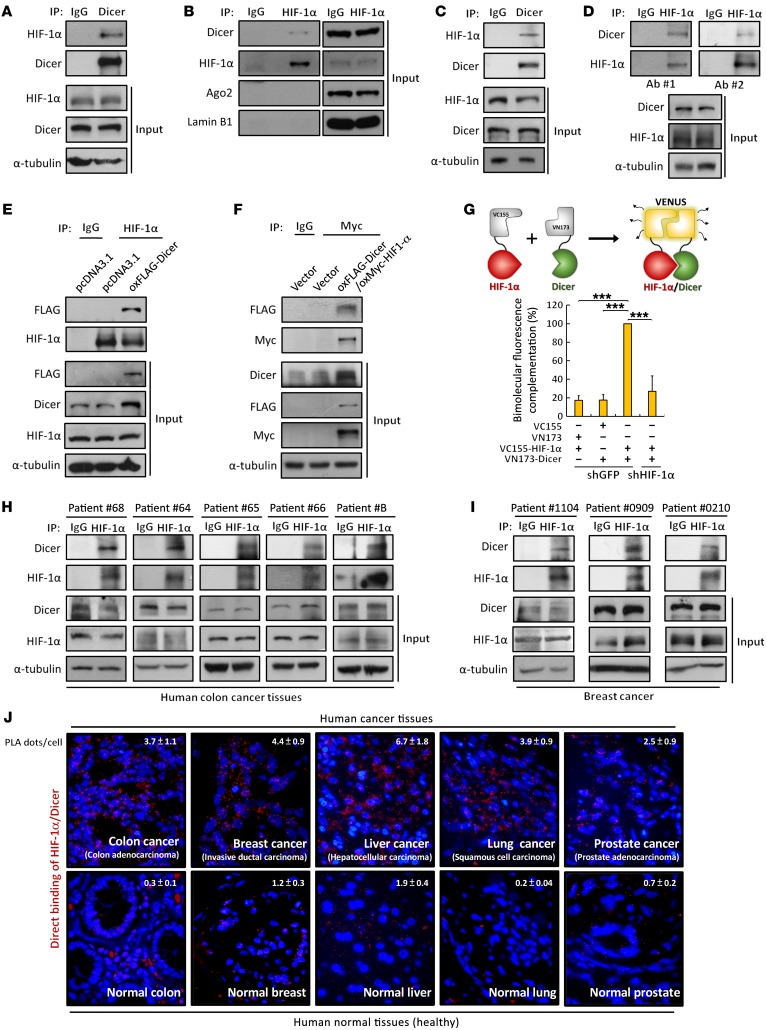
In our experiments originally intended to screen HIF-1α interaction proteins, we unexpectedly identified Dicer, the key biogenesis factor for miRNA maturation, as a HIF-1α–interacting protein through mass spectrometric (MS) analysis (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI89212DS1). To verify the association between HIF-1α and Dicer, we used anti-Dicer and anti–HIF-1α antibodies for immunoprecipitation and found that endogenous HIF-1α and Dicer interact with each other in HEK293T cells (Figure 1, A and B). Immunofluorescence staining also showed colocalization of HIF-1α and Dicer in the cytoplasm of HEK293T cells (Supplemental Figure 1C, arrowheads). Consistent results from immunoprecipitation were observed in HCT116 cells when using either anti-Dicer (Figure 1C) or 2 additional anti–HIF-1α antibodies (Figure 1D) to confirm their endogenous association. We further immunoprecipitated endogenous HIF-1α (Figure 1E) and exogenous Myc-tagged HIF-1α (Figure 1F) from the extracts of HCT116 cells transfected with FLAG-tagged Dicer and found that, similarly to endogenous Dicer, FLAG-Dicer was able to interact with either endogenous (Figure 1E) or exogenous Myc–HIF-1α (Figure 1F). Because HIF-1α is widely expressed among cancer cells, we collected a panel of cancer cell lines, including Caco-2, HT29, and SW480 (colon); MDA-MB-231 (breast); Mia-PaCa-2 (pancreas); A549 (lung); and HeLa (cervix); and determined the association between HIF-1α and Dicer. This interaction was broadly observed among all of these cancer cell lines from different tissue origins (Supplemental Figure 1D). We then performed bimolecular fluorescence complementation (BiFC) assay to study the direct protein interaction in living cells and found a significant reconstitution of Venus fluorescence signal in HCT116 cells expressing half of Venus-fused HIF-1α and Dicer, whereas knockdown of HIF-1α dramatically diminished the protein interaction (Figure 1G), confirming that HIF-1α specifically and directly interacts with Dicer. Furthermore, we utilized in situ proximity ligation assay (PLA) to investigate the endogenous binding of Dicer with HIF-1α (Supplemental Figure 1E). The PLA fluorescence signals derived from single protein-protein interaction events in close proximity (<40 nm) demonstrated significant direct interaction between HIF-1α and Dicer among all of the cancer cell lines used, as shown in Supplemental Figure 1D, whereas no signal was observed in any of the negative control cells (Supplemental Figure 1E). Because hypoxia induces HIF-1α nuclear localization, we isolated the subcellular fractions and performed immunoprecipitation to determine the interaction of HIF-1α/Dicer under hypoxic conditions. In the nuclear and cytoplasmic fractions isolated from HCT116 cells, immunoprecipitation showed that both nuclear and cytoplasmic HIF-1α accumulated under hypoxia (Supplemental Figure 1F, input), whereas the subcellular localization of Dicer was primarily restricted to the cytoplasm (Supplemental Figure 1F, input). Under hypoxia, the enhanced interaction of HIF-1α/Dicer was significantly observed in the cytoplasm (Supplemental Figure 1F, lane 4). Notably, in cell nucleus, HIF-1α/Dicer was induced under hypoxia, although the relative level was much lower than the interaction level in the cytoplasm (Supplemental Figure 1F, lane 2). Moreover, most of the HIF-1α/Dicer binding detected by in situ PLA was located in the cytoplasm, and few nuclear signals were observed under hypoxia (Supplemental Figure 1, G and H). These results suggested that HIF-1α primarily interacts with Dicer in the cytoplasm. Moreover, because a previous study has demonstrated that Dicer is also expressed in the nucleus (19), the HIF-1α/Dicer interaction may have another function in the nucleus and therefore require further investigation. To confirm the interaction between HIF-1α and Dicer in vivo, we collected surgically resected fresh tissues from colon (Figure 1H) and breast (Figure 1I) tumors and directly performed immunoprecipitation. Similarly to results from a series of cancer cell lines shown in Supplemental Figure 1, D and E, such binding was also observed in both colon and breast cancer tissues. We next used in situ PLA to analyze tissue array containing human normal and cancer tissues to investigate whether the direct binding could be identified in vivo, and consistently, direct interaction between HIF-1α and Dicer was clearly detected in colon, breast, lung, liver, and prostate cancer tissues, whereas fewer bindings were also observed in corresponding normal tissues (Figure 1J and Supplemental Figure 1I). These results indicate that HIF-1α associates with Dicer and that this interaction is a widespread phenomenon among different human cancer cell lines and tissues.
Figure 1. HIF-1α interacts with Dicer in multiple human cancer cell lines and tumors.
(A–D) Endogenous interaction between HIF-1α and Dicer. Immunoprecipitation of endogenous HIF-1α and Dicer were performed using anti–HIF-1α and anti-Dicer antibodies in HEK293T (A and B) and HCT116 cells (C and D). Ago2 was detected in anti–HIF-1α immunoprecipitates from HEK-293T cells. Lamin B1 was used as a loading control (B). (E and F) Immunoprecipitation analysis of the association between endogenous HIF-1α (E) or Myc–HIF-1α (F) and FLAG-Dicer in HCT116 cells. (G) BiFC of the association of VC115-fused HIF-1α and VN173-fused Dicer (top). HCT116 cells were transfected with VC155–HIF-1α, VN173-fused Dicer, or corresponding vector. (H–J) HIF-1α interacts with Dicer in tumor tissues. Anti–HIF-1α immunoprecipitates were isolated from lysates of surgically resected tumors to detect the in vivo interaction between HIF-1α and Dicer in colon (H) and breast (I) cancer tissues. The immunoblots presented were derived from replicate samples run on parallel gels (I). HIF-1α and Dicer were detected using in situ PLA in human colon, breast, lung, liver, and prostate normal and cancer tissues (J). PLA signals are shown in red along with DAPI nuclear staining (blue). Each red fluorescent dot indicates the direct binding of the HIF-1α/Dicer complex in close distance (<40 nm). Tissues stained with only anti–HIF-1α antibodies were also analyzed as negative controls shown in Supplemental Figure 1I. Data are presented as mean ± SD, with at least n = 3 per group. Multigroup comparisons were analyzed by 1-way ANOVA with Tukey’s post hoc test. ***P < 0.001.
HIF-1α downregulates Dicer protein expression under hypoxic and nonhypoxic conditions.
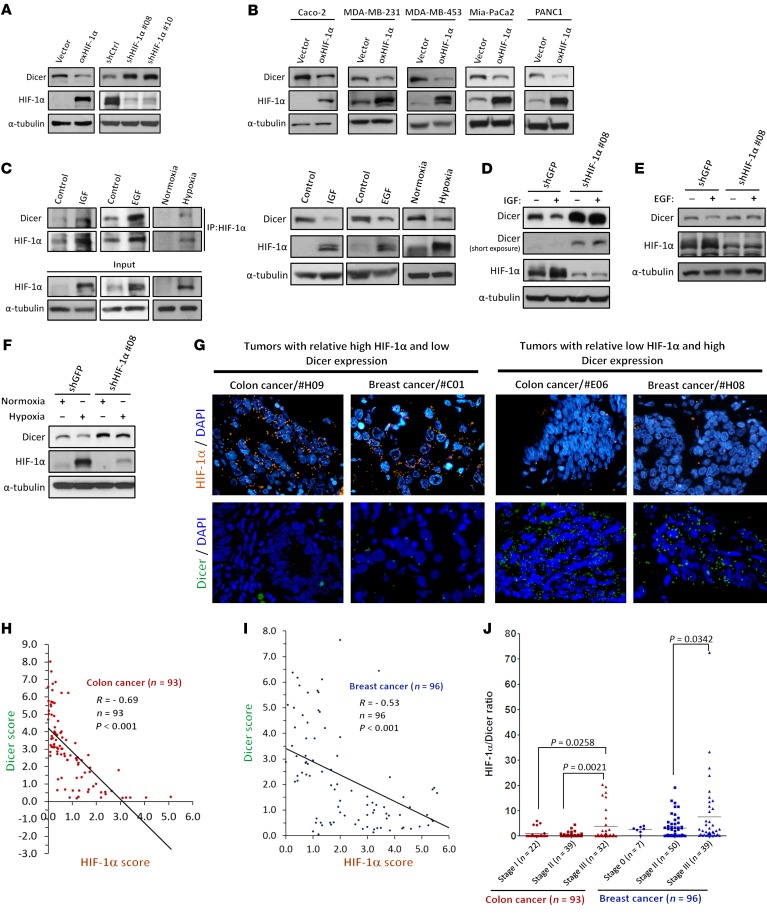
Having confirmed the ability of HIF-1α in binding to Dicer, we next investigated whether the expression level of Dicer was affected. We ectopically overexpressed HIF-1α and observed a significantly decreased protein level of Dicer, whereas knockdown of HIF-1α using 2 individual specific shRNAs dramatically enhanced Dicer protein expression (Figure 2A). However, the mRNA level of Dicer exhibited no significant change upon manipulation of HIF-1α (Supplemental Figure 2, A and B). The HIF-1α–mediated Dicer downregulation was further confirmed by restoration experiments in cells expressing shRNAs that target HIF-1α 3′ UTR, making the ectopically expressed HIF-1α insensitive to shRNA-mediated suppression (Supplemental Figure 2C, lanes 1–3). The downregulation was also observed in Caco-2, MDA-MB-231, MDA-MB-453, Mia-PaCa2, PANC1 cells overexpressing HIF-1α (Figure 2B), and in SW480, MDA-MB-231, Mia-PaCa2, and PANC1 cells with knockdown of HIF-1α (Supplemental Figure 2D), whereas Dicer mRNA levels remained unregulated (Supplemental Figure 2E). As a powerful oncogene, HIF-1α is induced under several pathophysiological circumstances, including growth factor stimulation and tumor hypoxia (1, 20). To investigate the regulation of Dicer in response to these HIF-1α–inducing microenvironmental factors, we treated cells with IGF and EGF or exposed them to hypoxia. Accordingly, both the binding between HIF-1α and Dicer and the downregulation (Figure 2C) of Dicer were observed in IGF- and EGF-treated HCT116 cells as well as in hypoxia-stressed cells. These effects of Dicer downregulation were further diminished in HIF-1α knockdown cells (Figure 2, D–F), suggesting an essential role of HIF-1α in the growth factor– and hypoxia-induced downregulation of Dicer. Again, the mRNA levels of Dicer were not altered upon growth factor stimulation (Supplemental Figure 2, F and G). Although the mRNA levels of Dicer were inhibited under hypoxia (Supplemental Figure 2H), knockdown of HIF-1α could not restore the repressed mRNA expression (Supplemental Figure 2I), suggesting that, unlike growth factor stimulation, hypoxia may suppress Dicer mRNA transcription through a HIF-1α–independent manner, which was described previously (21–23). The above experiments involving genetic manipulations and biological stimulations suggest that HIF-1α posttranscriptionally downregulates Dicer protein expression.
Figure 2. HIF-1α downregulates Dicer protein expression.
(A) Effects of HIF-1α on the expression of Dicer. The expression of Dicer was determined in HCT116 cells in which HIF-1α was overexpressed or knocked down by 2 specific shRNAs. The immunoblots presented were derived from replicate samples run on parallel gels. oxHIF-1α, overexpressed HIF-1α. (B) HIF-1α suppresses the protein expression of Dicer in multiple cancer cell lines. The cell lysates from Caco-2, MDA-MB-231, MDA-MB-453, Mia-PaCa-2, or PANC-1 with HIF-1α overexpression were subjected to Western blot analysis. The immunoblots presented were derived from replicate samples run on parallel gels (Caco-2). (C) HCT116 cells were treated with IGF or EGF and exposed to hypoxic conditions. The binding of Dicer with HIF-1α was determined through immunoprecipitation (left). (D–F) Similar experiments were performed in shHIF-1α–expressing HCT116 cells to study the functional roles of HIF-1α. The immunoblots presented were derived from replicate samples run on parallel gels (D). (G) Representative colon and breast cancer tissues showing the expression of either combined high HIF-1α and low Dicer or low HIF-1α and high Dicer. Each dot of fluorescence signal was generated from 2 closely contacted antibodies recognizing different epitopes of the same target protein (HIF-1α, orange; Dicer, green). Original magnification: ×600. (H and I) Dicer and HIF-1α scores were counted and calculated as PLA dots/cell for analyzing their expression correlation in colon (H) and breast (I) cancers. (J) The HIF-1α/Dicer ratio was used to study clinical associations with stage in both colon and breast cancers. Data are presented as mean ± SD, with at least n = 3 per group. Unpaired, independent groups of 2 were analyzed by 2-tailed Student’s t test.
In line with our previous results showing that HIF-1α interacts and downregulates Dicer protein expression, we further investigated whether higher levels of HIF-1α protein correlate with repressed Dicer protein levels in human cancer tissues. We first used 2 specific antibodies recognizing different epitopes on the same protein to perform in situ PLA for quantitatively determining protein expression with improved specificity, as the florescence signals were only detected when 2 antibodies simultaneously bound to the same target protein (Figure 2G). Consistent with our in vitro results, protein expression of HIF-1α was inversely correlated with Dicer protein levels in 93 human colon cancer tissues (Figure 2H; R = –0.69, P < 0.001) and 96 breast cancer tissues (Figure 2I; R = –0.53, P < 0.001), supporting the HIF-1α–mediated downregulation of Dicer in both cancer types. We further analyzed the ratio of HIF-1α to Dicer for reflecting levels of HIF-1α–suppressed Dicer expression, as greater ratios indicate patients have simultaneously higher HIF-1α and lower Dicer in the tumors and vice versa. Interestingly, the HIF-1α/Dicer ratio was significantly higher in stage 3 tumors compared with those from either stage 1 or 2 in colon cancer (Figure 2J; P = 0.0258 and P = 0.0021, respectively), and similarly, the ratio was significantly higher in stage 3 tumors compared with stage 2 in breast cancer (Figure 2J; P = 0.0342). But no statistically significant association with T stages was found in both cancer types (Supplemental Figure 2J). In addition, the HIF-1α/Dicer ratio was significantly higher in tumors from colon (Supplemental Figure 2K, left; P = 0.004) and breast (Supplemental Figure 2K, right; P = 0.007) cancer patients with lymph node metastasis. This clinical evidence supports HIF-1α–mediated Dicer downregulation in cancer, and this regulation significantly associates with advanced stages and the presence of lymph node metastasis in human colon and breast cancer patients.
HIF-1α nontranscriptionally facilitates autophagy-mediated proteolysis of Dicer.
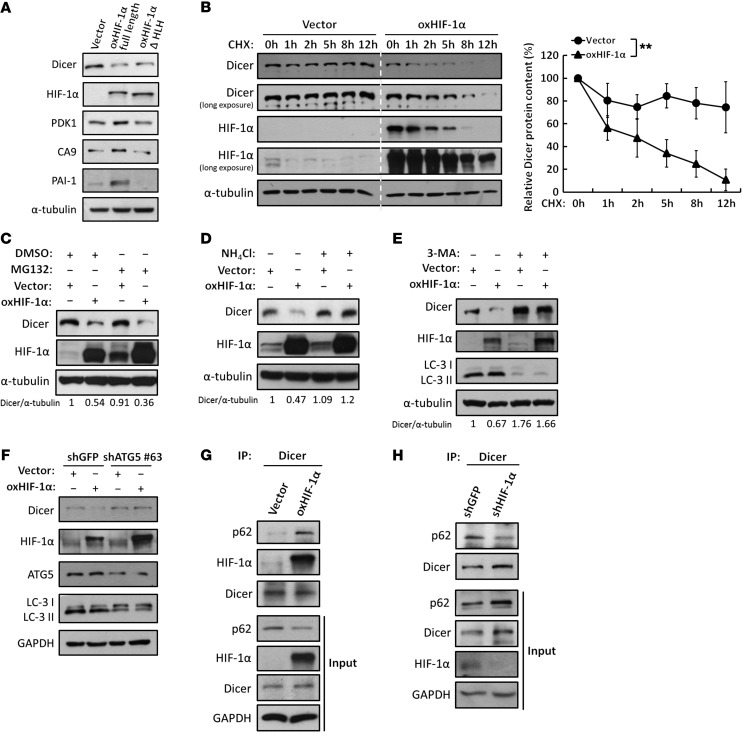
HIF-1α has been studied for decades regarding its transcriptional activity, which was previously shown to regulate gene expression through the recognition of a HIF-1α response element by a helix-loop-helix (HLH) DNA-binding motif. To investigate whether the transcriptional activity of HIF-1α is required for the regulation of Dicer, we compared the abilities of HLH-truncated (ΔHLH) and full-length HIF-1α to downregulate Dicer. Interestingly, the downregulation of Dicer was unaffected by removal of the DNA-binding ability of HIF-1α. In both overexpression and restoration experiments, HLH-truncated HIF-1α exhibited effects similar to those of full-length HIF-1α in suppressing Dicer, whereas the well-known transcriptional targets PDK1, CA9, and PAI-1 were induced by only full-length HIF-1α in HCT116 and HEK293T cells (Figure 3A, Supplemental Figure 2C, lanes 2 and 4, and Supplemental Figure 3A). These results suggest a transcription-independent role of HIF-1α in regulating Dicer. To investigate the underlying mechanism leading to repressed Dicer protein expression, we determined its protein stability in HCT116 and Mia-PaCa2 cells overexpressing HIF-1α. In the presence of cycloheximide (CHX), Dicer protein expression was stably maintained for 12 hours, whereas a significant decrease was observed at 2 to 8 hours in HIF-1α–overexpressing HCT116 and Mia-PaCa2 cells (Figure 3B and Supplemental Figure 3B), indicating that HIF-1α accelerates the degradation of Dicer protein. Similar effects were also observed in IGF-treated cells, as knockdown of HIF-1α further abolished the IGF-mediated Dicer degradation, suggesting that IGF induces HIF-1α expression to destabilize Dicer protein (Supplemental Figure 3C). Mammalian cells use proteasome and lysosome systems as major proteolytic pathways. Thus, we first determined whether this pathway acts in a proteasome-dependent manner. However, treatment with the proteasome inhibitor MG132 could not prevent the reduction of Dicer expression in HIF-1α–overexpressing cells (Figure 3C), suggesting that HIF-1α promotes proteasome-independent degradation of Dicer. These results prompted us to investigate the involvement of the lysosomal pathway. Because the expression of Dicer was restored in HIF-1α–overexpressing cells in the presence of the lysosomal inhibitor NH4Cl (Figure 3D), it can be concluded that lysosome-mediated proteolysis is involved in the HIF-1α–enhanced degradation of Dicer. These findings led us to further investigate the role of autophagy, a pathway that selectively delivers proteins to the lysosome for further degradation (24). We treated cells with 3-methyladenine (3-MA) and observed that inhibiting autophagy completely prevented the HIF-1α–mediated degradation of Dicer (Figure 3E). Similar results were also obtained when a specific shRNA targeting autophagy protein 5 (ATG5) was used to prevent autophagy (Figure 3F), which further confirmed the role of an autophagy-dependent pathway for the degradation of Dicer. To determine whether Dicer is recruited into autophagosomes as a cargo protein, we evaluated the level of Dicer recognized by p62, an autophagy receptor that is known to form complexes with the cargo proteins (25). The coimmunoprecipitation results showed that the p62-Dicer complex was increased or decreased in HIF-1α–overexpressing or HIF-1α–knockdown HCT116 cells, respectively (Figure 3, G and H). Additionally, Dicer translocated into autophagosomes isolated from HIF-1α–overexpressing cells (Supplemental Figure 3D), and supportively, confocal images of immunofluorescence staining showed the increased colocalization of Dicer/p62 in the cytoplasm (Supplemental Figure 3E). These results demonstrate that HIF-1α facilitates the recognition of Dicer by autophagosomes for further degradation.
Figure 3. HIF-1α enhances proteolysis of Dicer through an autophagy-lysosomal pathway.
(A) Effects of the nontranscriptional activity of HIF-1α on Dicer. WT HIF-1α or HLH-truncated HIF-1α was overexpressed in HCT116 cells for Western blot analysis to determine the expression of Dicer, PDK1, CA9, and PAI-1. The immunoblots presented were derived from replicate samples run on parallel gels. (B) HIF-1α enhances the protein degradation of Dicer. Cells were treated with CHX at the indicated times to block de novo protein synthesis. The expression of Dicer was determined by Western blot analysis and was further quantified by ImageJ (NIH). The relative levels of Dicer protein remaining at each time point were normalized to α-tubulin (right). Data are presented as mean ± SD, with at least n = 3 per group. **P < 0.01, 2-way ANOVA. All of the cell lysates were run on the same gel. (C–E) Effects of the proteolytic pathways on the HIF-1α–induced downregulation of Dicer. Cells were treated with MG132 to inhibit proteasomal degradation (C), ammonium chloride (NH4Cl) to inhibit lysosomal degradation (D), or 3-MA to inhibit autophagic degradation (E). The immunoblots presented were derived from replicate samples run on parallel gels. The ratios of Dicer/α-tubulin protein levels were quantified by using ImageJ. (F) Similar experiments were performed in cells in which ATG5 was genetically knocked down to inhibit autophagy. LC3 was detected for autophagic activation. (G and H) The associations between Dicer and the autophagy receptor p62 were analyzed in the presence of NH4Cl and CQ to block autophagy-lysosomal degradation in HIF-1α–overexpressing (G) or HIF-1α knockdown (H) HCT116 cells. The immunoblots presented were derived from replicate samples run on parallel gels (H).
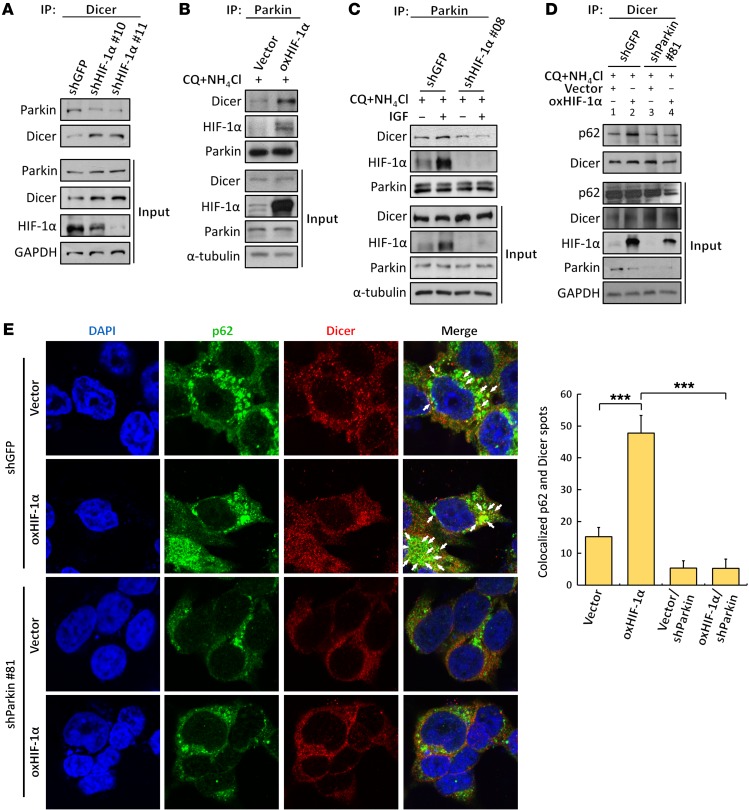
HIF-1α recruits Parkin E3 ubiquitin ligase, which ubiquitinates Dicer for recognition by autophagosomes.
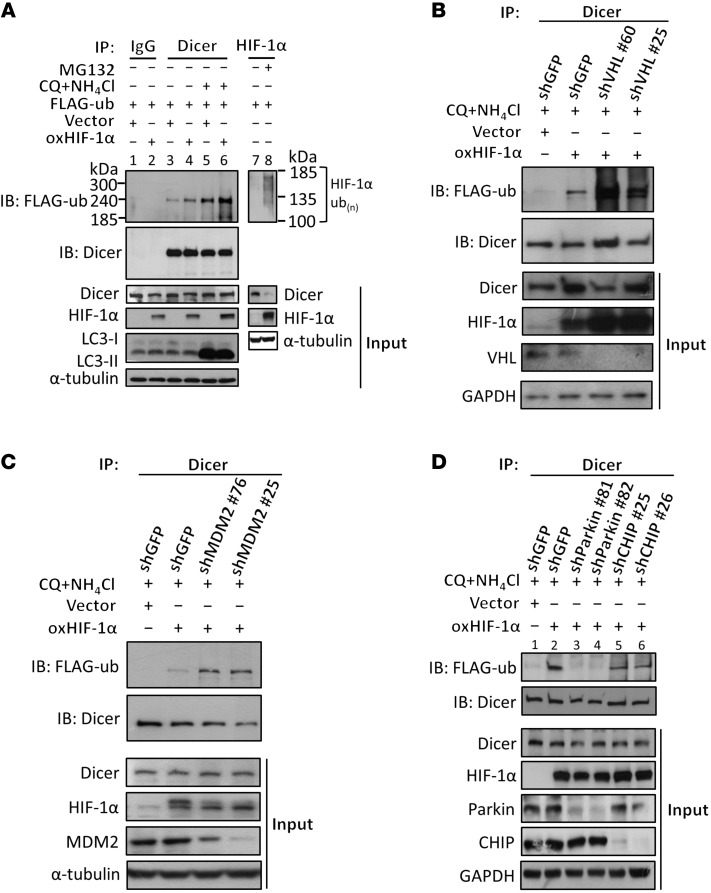
We next sought to investigate an interesting question of how Dicer is recognized by the autophagy pathway. Recent evidence has shown that the ubiquitination of cargo proteins is a selective mechanism through which the substrate is recognized by the autophagy receptor (25–27). Hence, we determined the ubiquitination level of Dicer and observed an obvious induction of ubiquitinated Dicer in HIF-1α–overexpressing HEK293T and HCT116 cells either in the absence or presence of lysosomal inhibitors (Figure 4A, lanes 4 and 6, and Supplemental Figure 4A, lanes 4 and 6). In addition, the ubiquitination was significantly induced by IGF stimulation (Supplemental Figure 4B, lane 4), whereas knockdown of HIF-1α completely abolished this effect (Supplemental Figure 4B, lane 6). These results suggest that Dicer ubiquitination is enhanced either by ectopically expressed or by IGF-induced HIF-1α, which prompted us to identify the E3 ligase responsible for catalyzing Dicer ubiquitination. NH4Cl and chloroquine (CQ) were used to block the HIF-1α–induced autolysosomal degradation of the experiments shown in Figure 4. Thus, the protein ubiquitination or level of protein interaction was clearly determined. We first hypothesized that, through a protein-protein interaction, HIF-1α may recruit VHL or MDM2 to facilitate the ubiquitination of Dicer, since they are the known E3 ligases physically associated with HIF-1α (28, 29). However, HIF-1α–induced ubiquitination was not abolished by knockdown of either VHL or MDM2 (Figure 4, B and C). Both experiments showed that neither VHL nor MDM2 is essential for Dicer ubiquitination. In VHL-knockdown cells, the enhanced ubiquitination might have indirectly resulted from HIF-1α accumulation (input), thus increasing formation of HIF-1α/E3 ligase/Dicer complexes (the E3 ligase was thereafter identified as Parkin). In MDM2-knockdown cells, the enhanced ubiquitination might have reflected another unknown pathway instead of resulting from increased HIF-1α expression (HIF-1α is not upregulated in input shown in Figure 4C). Thus, we excluded the functional roles of these proteins in this mechanism. CHIP and Parkin are 2 of the major E3 ligases involved in ubiquitinating substrate proteins for the recognition of autophagy pathway (24, 30–32). Thus, we next knocked down the expression of CHIP and Parkin by shRNAs and found that the HIF-1α–mediated Dicer ubiquitination was completely abolished by suppressing Parkin (Figure 4D, lanes 3 and 4), whereas knockdown of CHIP had only slight effects (Figure 4D, lanes 5 and 6). Similarly, ectopically expressed Parkin enhanced the HIF-1α–induced ubiquitination of Dicer (Supplemental Figure 4C, lane 5), whereas no effects were observed in HCT116 cells overexpressing ParkinC431S in which the putative catalytic cysteine had been mutated to serine (Supplemental Figure 4C, lane 6), indicating that Parkin is required for Dicer ubiquitination. Moreover, we knocked down Parkin to confirm its functional role in downregulating Dicer and found that both HIF-1α– and IGF-suppressed Dicer expression were abolished (Supplemental Figure 4, D and E). We next investigated the role of HIF-1α in recruiting Parkin to form complex with Dicer. NH4Cl and CQ were used to block the HIF-1α–induced autolysosomal degradation of Dicer in most of the experiments shown in Figure 5, except Figure 5A. Thus, the protein ubiquitination or level of protein interaction was clearly determined. We immunoprecipitated Parkin and observed a significantly decreased level of coimmunoprecipitated Dicer upon HIF-1α knockdown (Figure 5A) and vice versa (Figure 5B). In addition, HIF-1α brought Parkin and Dicer together in response to IGF treatment, as knockdown of HIF-1α prevented Dicer/Parkin complex formation facilitated by IGF (Figure 5C), which further supports that HIF-1α is a bridge required for the interaction between Parkin and Dicer. These results demonstrate a dynamic mechanism that HIF-1α recruits Parkin to associate with Dicer for ubiquitination, thus promoting the recognition of ubiquitinated Dicer by autophagy receptor, which was also confirmed by immunoprecipitation (Figure 5D, lanes 2 and 4) and immunofluorescence analysis (Figure 5E), showing that knockdown of Parkin obviously diminished the HIF-1α–induced association of Dicer with p62.
Figure 4. HIF-1α induces ubiquitination of Dicer.
(A) FLAG-ubiquitin was expressed in HEK293T cells for Dicer ubiquitination assays. The immunoprecipitates isolated by anti-Dicer antibodies were subjected to Western blot analysis for determining the expression of FLAG-ubiquitin, indicating the ubiquitination levels of Dicer. LC3 was detected for autophagosome accumulation after treatment with CQ and NH4Cl. Cells were treated with MG132 to enhance the accumulation of poly-ubiquitinated HIF-1α as a positive control (lane 8). (B–D) Effects of different E3 ubiquitin ligases on HIF-1α–induced Dicer ubiquitination. VHL (B), MDM2 (C), Parkin, and CHIP (D) were knocked down in HIF-1α–expressing HCT116 cells, and the level of Dicer ubiquitination was measured. The experiments in B–D were performed in the presence of CQ and NH4Cl to block autophagy-lysosomal degradation.
Figure 5. HIF-1α recruits Parkin to interact with Dicer for autophagic proteolysis.
(A–C) Roles of HIF-1α in the recruitment of Parkin to Dicer. Associations between Dicer and Parkin in HCT116/shHIF-1α (A), HCT116/HIF-1α (B), and IGF-treated HCT116 (C) cells were detected. The immunoblots presented were derived from replicate samples run on parallel gels (B). (D and E) Effects of Parkin knockdown on the interaction between p62 and Dicer. Immunoprecipitates isolated by anti-Dicer antibodies were harvested to detect the protein association with p62 (D). The immunoblots presented were derived from replicate samples run on parallel gels. (E) Immunofluorescence analysis was performed using anti-p62, anti-Dicer antibodies, and DAPI for nuclear staining. Images were obtained by using confocal microscopy, as indicated: Dicer (red); p62 (green); DAPI (blue) (E, left). The spots of p62 and Dicer were quantified by using ImageJ (E, right). The experiments in B–E were performed in the presence of CQ and NH4Cl to block autophagy-lysosomal degradation. Original magnification: × 1000. Data are presented as mean ± SD, with at least n = 3 per group. Multigroup comparisons were analyzed by 1-way ANOVA with Tukey’s post hoc test. ***P < 0.001.
Identification of functional domains required for reciprocal protein interaction.
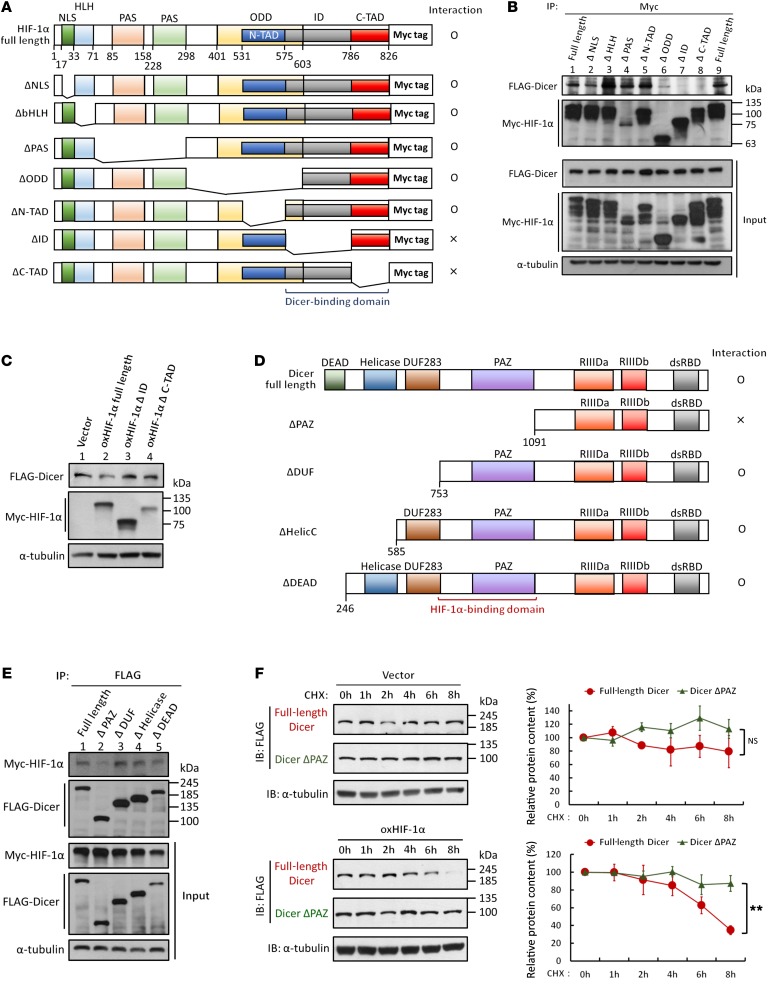
To further investigate which domain of HIF-1α is responsible for binding to Dicer, we generated 7 deletion mutants (ΔNLS, ΔHLH, ΔPAS, ΔODD, ΔN-TAD, ΔID, ΔC-TAD) of Myc–HIF-1α in which individual functional domains were truncated (Figure 6A). These truncation mutants were cotransfected with FLAG-Dicer in HEK293T and HCT116 cells to investigate their interaction. The coimmunoprecipitation results showed that deletions of HIF-1α from NLS to N-TAD did not reduce the binding between HIF-1α and Dicer (Figure 6B, lanes 2–5; Supplemental Figure 5, lanes 2–5), whereas truncating the ID and C-TAD domains dramatically disrupted the interaction with Dicer (Figure 6B, lanes 7 and 8; Supplemental Figure 5, lanes 7 and 8). It was also noted that deleting the ODD domain partially reduced the association (Figure 6B, lane 6). However, as the ODD domain contains both a complete N-TAD domain, which is not dispensable for binding (Figure 6, A and B), and part of the ID domain, which is required for binding (Figure 6, A and B), the reduction of the protein interaction observed in the ΔODD group may result from the partial deletion of ID domain in ODD-truncated HIF-1α. Because HLH and N-TAD domains are important regions for HIF-1β and prolyl hydroxylase domain protein (PHD) binding (33–36), truncating these domains disrupts the association between HIF-1β or PHD and HIF-1α. In line with the results showing the enhanced interaction between ΔHLH or ΔN-TAD HIF-1α and Dicer, it is likely that the binding of HIF-1β or PHD may repress the interaction of HIF-1α with Dicer. Because the ΔID and ΔC-TAD truncations exhibited greatly reduced interaction with Dicer, we also examined their functions and consistently found that these 2 mutants exhibited no regulatory ability on downregulating Dicer protein expression (Figure 6C, lanes 3 and 4). On the other hand, we used 4 serially truncated Dicer mutants (ΔPAZ, ΔDUF, ΔHelicC, ΔDEAD) to identify the region required for the interaction with HIF-1α (Figure 6D). The FLAG-Dicer truncations were coexpressed with Myc–HIF-1α in cells for further coimmunoprecipitation assays. The ΔDUF283, ΔHelicase, and ΔDEAD mutants of Dicer retained a strong interaction with HIF-1α (Figure 6E, lanes 3 and 5), whereas deletion of PAZ significantly disrupted this interaction (Figure 6E, lane 2). Also, this PAZ-deleted Dicer was observed to be insensitive to HIF-1α–induced protein degradation (Figure 6F; PAZ-deleted Dicer in HIF-1α–overexpressing cells). These results suggest that the ID to C-TAD domain of HIF-1α and the PAZ domain of Dicer are essential for their functional protein-protein interaction.
Figure 6. Identification of functional domains required for HIF-1α–Dicer interaction.
(A) Schematic representation of the Myc-tagged truncated HIF-1α used in this study. The immunoblots presented were derived from replicate samples run on parallel gels. (B) FLAG-Dicer and individual Myc-tagged HIF-1α truncations were coexpressed in HCT116 cells. The immunoprecipitates isolated by anti–Myc-tag antibodies were subjected to Western blot analysis to detect the interaction between truncated Myc–HIF-1α and FLAG-Dicer. (C) Effects of HIF-1α with a truncated ID or C-TAD domain on the downregulation of Dicer. Lysates from HCT116 cells expressing full-length HIF-1α or the ΔID or ΔC-TAD truncations were subjected to Western blot analysis. The immunoblots presented were derived from replicate samples run on parallel gels. (D) Schematic representation of the FLAG-tagged truncated Dicer used in this study. (E) Myc–HIF-1α and individual FLAG-tagged Dicer truncations were coexpressed in HCT116 cells. Immunoprecipitates isolated by anti–FLAG-tag antibodies were subjected to Western blot to determine the interaction between truncated Dicer and Myc–HIF-1α. (F) PAZ domain–deleted Dicer was insensitive to HIF-1α–mediated protein degradation. HCT116 cells were transfected with full-length or PAZ truncation of Dicer, and protein stability was monitored in the presence of CHX at the indicated times to block de novo protein synthesis. Data are presented as mean ± SD, with at least n = 3 per group. **P < 0.01, 2-way ANOVA. The experiments depicted in B and E were performed in the presence of CQ and NH4Cl to block autophagy-lysosomal degradation.
Downregulation of Dicer decreases processing efficiency and diminishes target inhibition by miRNAs.
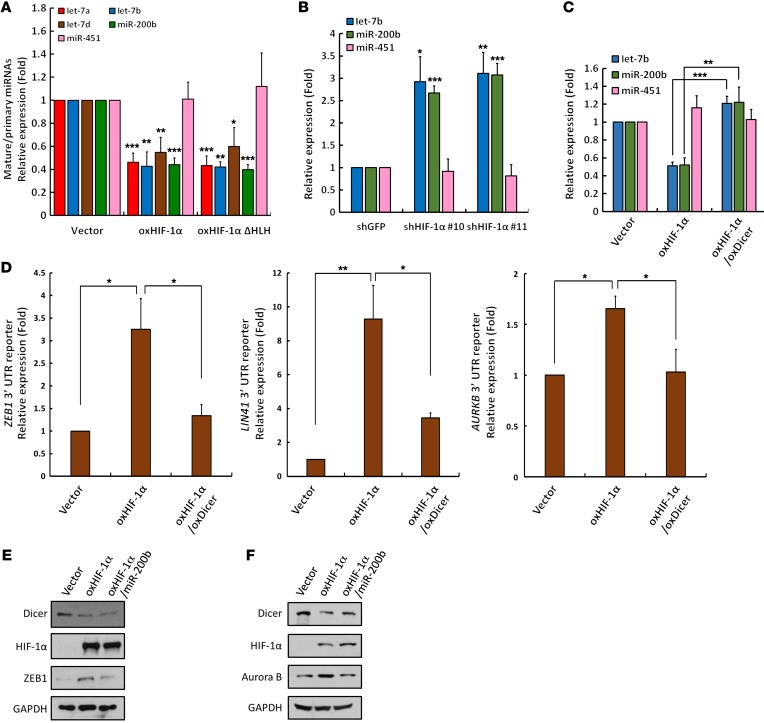
To validate the consequences of Dicer suppression, we determined the expression of let-7 family miRNAs and miR-200b, which are highly conserved miRNAs known to be sensitive to Dicer processing (15, 16, 37); miR-451, which is generated through a Dicer-independent pathway (38); and miR-210, which is a known transcriptional target downstream of HIF-1α (39, 40) (Supplemental Figure 6A). We simultaneously measured the expression of their precursors (pri-miRNAs; Supplemental Figure 6B) to determine the ratio of mature to pri-miRNAs as an indicator of biogenesis activity (Figure 7A). Both pri- and mature miR-210 were upregulated in cells overexpressing WT, but not ΔHLH-truncated HIF-1α (Supplemental Figure 6, A and B), which confirmed that the transcriptional activity of HIF-1α is essential for the regulation of miR-210. The expression of let-7 family miRNAs and miR-200b was significantly reduced in cells transfected with either WT or ΔHLH-truncated HIF-1α, whereas the level of miR-451, a miRNA generated from the Ago-2–mediated Dicer-independent pathway, remained unchanged in these experiments (Figure 7A and Supplemental Figure 6, A and B), suggesting that HIF-1α posttranscriptionally suppresses the processing activity of Dicer. Similar results from the knockdown of HIF-1α confirmed this regulation, as the levels of let-7b and miR-200b were enhanced, but miR-451 was unaffected (Figure 7B). In line with this evidence, let-7b and miR-200b levels were reconstituted in Dicer-restored cells overexpressing HIF-1α, indicating that HIF-1α–mediated Dicer repression results in downregulatuion of the miRNAs (Figure 7C). The let-7 family miRNAs and miR-200b are well-studied tumor suppressors, and their downregulation promotes malignant phenotypes of cancer cells, including facilitating the epithelial-mesenchymal transition (EMT) (41, 42). Our clinical results also supported that HIF-1α–suppressed Dicer expression significantly correlates with lymph node metastasis in cancer patients (Supplemental Figure 2K). To evaluate the biological activities of these miRNAs, we analyzed their effects on suppressing the expression of luciferase genes harboring the 3′ UTRs of ZEB1 (target of miR-200b) (41), LIN41 (target of let-7) (43), and Aurora B (AURKB) (target of let-7) (44). The expression of all of these reporters was significantly enhanced upon HIF-1α overexpression, whereas restoring Dicer abolished these effects (Figure 7D). Moreover, Western blot analysis also showed that restoring miR-200b or let-7 suppressed expression of ZEB1 or Aurora B induced by HIF-1α, respectively (Figure 7, E and F). These results suggest that HIF-1α–induced Dicer degradation suppresses miRNA maturation and consequently decreases the activity of Dicer-dependent miRNAs.
Figure 7. HIF-1α–enhanced Dicer proteolysis in miRNA biogenesis.
(A–C) Effects of HIF-1α on Dicer-dependent miRNA maturation. HCT116 cells overexpressing HIF-1α or HLH domain–truncated HIF-1α (A), knockdown of HIF-1α (B), or restoration of Dicer (C) were subjected to quantitative reverse-transcription PCR (qRT-PCR) for analyzing the expression of miRNAs, including let-7 family members (let-7a, let-7b, let-7d), miR-200b, and miR-451. (A) The ratios of mature to pri-miRNAs for let-7a, let-7b, let-7d, miR-200b, and miR-451 were determined using data sets shown in Supplemental Figure 6, A and B. (D–F) Effects of HIF-1α–regulated Dicer on the expression of miR-200b and let-7b–targeted genes. HCT116 cells were transfected with ZEB1, LIN41, or AURKB 3′ UTR luciferase reporter. The expression of luciferase was assayed in HIF-1α–overexpressing cells with or without Dicer restoration (D). miR-200b (E) or let-7b (F) was restored in HIF-1α–expressing cells. Aurora B, let-7b–targeted gene was analyzed by Western blot. The immunoblots presented were derived from replicate samples run on parallel gels (E). Data are presented as mean ± SD, with at least n = 3 per group. Unpaired, independent groups of 2 were analyzed by 2-tailed Student’s t test. Multigroup comparisons were analyzed by 1-way ANOVA with Tukey’s post hoc test.
HIF-1α–induced Dicer degradation facilitates EMT and cancer metastasis through the suppression of miR-200b maturation.
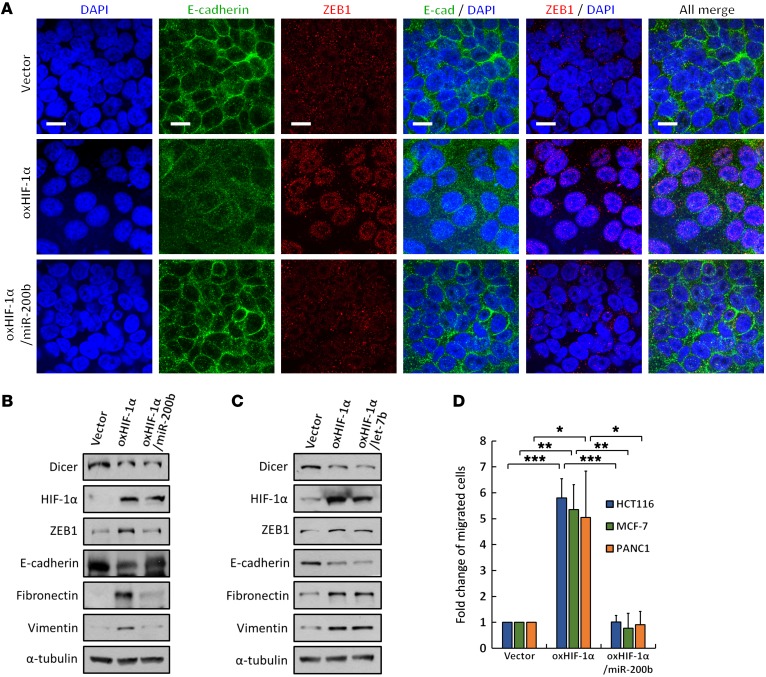
To explore the biological consequences of HIF-1α–suppressed miR-200b expression, we analyzed the protein distribution and expression of EMT markers in HCT116 cells. ZEB1-mediated EMT was observed in HIF-1α–overexpressing cells, including the disruption of intercellular E-cadherin junctions, enhancement of nuclear ZEB1 translocation (Figure 8A), upregulation of the mesenchymal markers fibronectin and vimentin, and downregulation of E-cadherin (Figure 8B). These effects were reconstituted in miR-200b–coexpressing cells. However, the restoration of let-7b did not suppress HIF-1α–induced EMT (Figure 8C), indicating a functional role of HIF-1α–suppressed miR-200b in regulating EMT. The resulting enhanced migratory ability was also consistently observed in HIF-1α–overexpressing HCT116 (colon), MCF-7 (breast), and PANC1 (pancreas) cells and was further diminished upon the restoration of miR-200b (Figure 8D). Furthermore, ΔHLH HIF-1α was overexpressed in different cell types to study its transcriptional-independent roles. Immunofluorescence staining demonstrated that E-cadherin expression was also repressed, whereas ZEB1 accumulated in the nucleus of ΔHLH HIF-1α–expressing HCT116, MCF-7, and PANC1 cells (Supplemental Figure 7A). In agreement with previous results, fibronectin and vimentin were induced along with E-cadherin repression in ΔHLH HIF-1α–expressing cells. These effects were further diminished by the restoration of miR-200b, but Aurora B protein remained enhanced after the restoration of miR-200b (Supplemental Figure 7B). The migration ability of ΔHLH HIF-1α–expressing cancer cells was enhanced, whereas the restoration of miR-200b diminished this effect (Supplemental Figure 7C). These results indicated that HIF-1α–induced Dicer degradation facilitates the EMT of cancer cells through the suppression of miR-200b maturation.
Figure 8. Functional roles of HIF-1α–induced miR-200b suppression in EMT.
(A–D) Roles of HIF-1α in miR-200b and let-7b–mediated EMT and cell migration. HIF-1α and miR-200b or let-7b were coexpressed in HCT116 cells. ZEB1 and E-cadherin were analyzed through immunofluorescence staining (A) using anti–E-cadherin and anti-ZEB1 antibodies or Western blotting (B and C) and using anti–E-cadherin, anti-ZEB1, anti-fibronectin, and anti-vimentin antibodies. Immunofluorescence images were obtained by using confocal microscopy, as indicated: E-cadherin (green); ZEB1 (red); DAPI (blue). Scale bars: 10 μm. The effects on cell migration in HCT116, MCF-7, and PANC1 cells were measured in Boyden chamber assays (D). The immunoblots presented were derived from replicate samples run on parallel gels (B and C). Data are presented as mean ± SD, with at least n = 3 per group. Multigroup comparisons were analyzed by 1-way ANOVA with Tukey’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001.
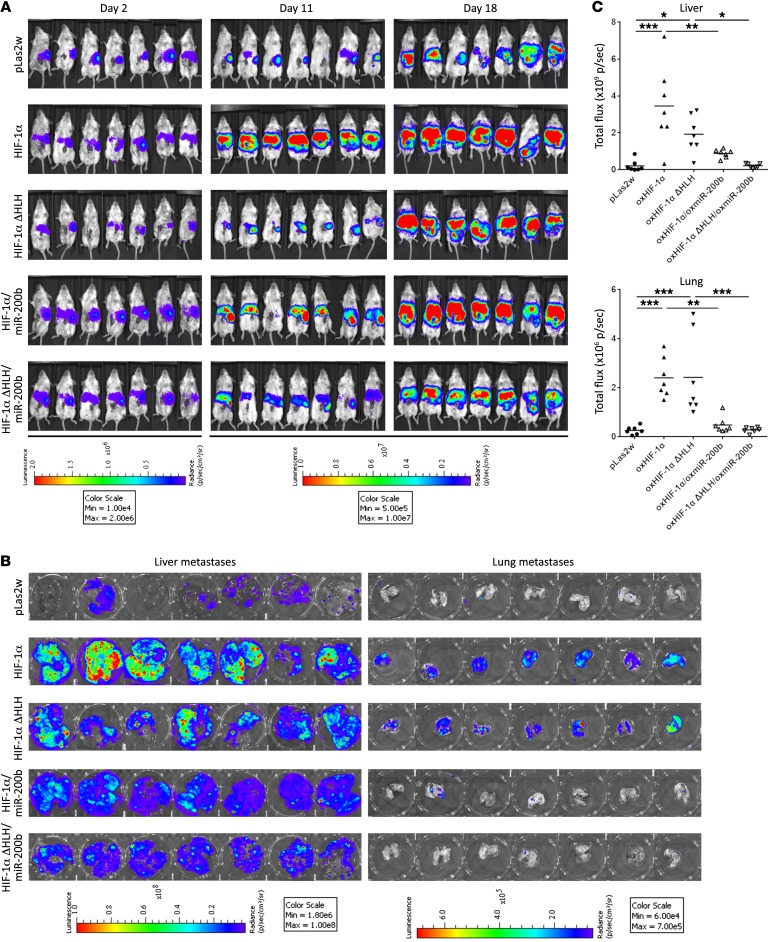
To evaluate the metastatic effects in vivo, we performed intrasplenic injection (45, 46) to determine spontaneous metastasis. This model enables the spontaneous metastasis of the primary tumor through the portal system, thus mimicking the metastatic pattern in human colon cancer and generating liver and lung metastasis (47–50). miR-200b was restored in CT-26/Luc cells stably expressing either WT or HLH-truncated HIF-1α for metastasis in animal experiments. To confirm that CT-26 cells were equally injected into the mouse spleen, the photon intensities of the primary sites were measured at 2 days after injection (Figure 9A and Supplemental Figure 7D). At 11 days after splenic injection, the intensities of the metastatic signals in the liver were observed in CT-26 cells expressing either WT or HLH-truncated HIF-1α (Figure 9A). At 18 days after splenic injection, the livers and lungs were isolated and the photon intensities were detected. The intensities of the metastatic signals in the liver and lungs were strongly enhanced in CT-26 cells expressing either WT or HLH-truncated HIF-1α, whereas the enhanced metastasis was further diminished by restoring miR-200b, because WT or HLH-truncated HIF-1α–induced liver and lung metastasis were significantly abolished in cells overexpressing miR-200b (Figure 9, B and C). Our in vivo evidence further supported the in vitro results suggesting that HIF-1α downregulates Dicer, thereby suppressing miR-200b maturation and consequently facilitating cancer metastasis.
Figure 9. HIF-1α–induced miR-200b suppression facilitates cancer metastasis.
(A–C) CT-26/Luc mouse colon cancer cells stably expressing WT or HLH-truncated HIF-1α were restored with miR-200b for metastasis animal experiments (A). After intrasplenic injection, tumor metastasis was monitored by in vivo images of mouse (A) liver and lung (B). (C) The photon intensities of the metastatic signals in the livers and lungs were calculated 18 days after intrasplenic injection (n = 7 per group). Data are presented as mean ± SD, with at least n = 3 per group. Multigroup comparisons were analyzed by 1-way ANOVA with Tukey’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
The dysregulation of miRNAs has been implicated in human diseases. Here, we demonstrate a mechanism by which HIF-1α selectively enhances the ubiquitination and autophagic proteolysis of Dicer and hinders the biogenesis of miRNA, thus leading to aberrant miRNA maturation and enhanced EMT in human cancers (Figure 10). Recent evidence has suggested that, beyond the current knowledge of the “canonical” pathway for generating miRNAs, several cancer-related protein factors, such as p53, BRCA1, and Smad, participate in the modulation of miRNA biogenesis reviewed previously (10, 12). Because these pathways can independently or coordinately orchestrate signaling networks and regulate cell function, the reciprocal regulation with HIF-1α requires further investigation. For example, the crosstalk between HIF-1α and p53 pathways regulates tumor growth at different stages of cancer progression (51). Therefore, in contrast with tumor-suppressive p53, which interacts with the Drosha complex and enhances the processing of primary miRNA (52), the oncogenic HIF-1α acts as a gatekeeper for cancer cells, thus preventing the maturation of pre-miRNA by Dicer and resulting in fine-tuning the balance modulating cancer progression.
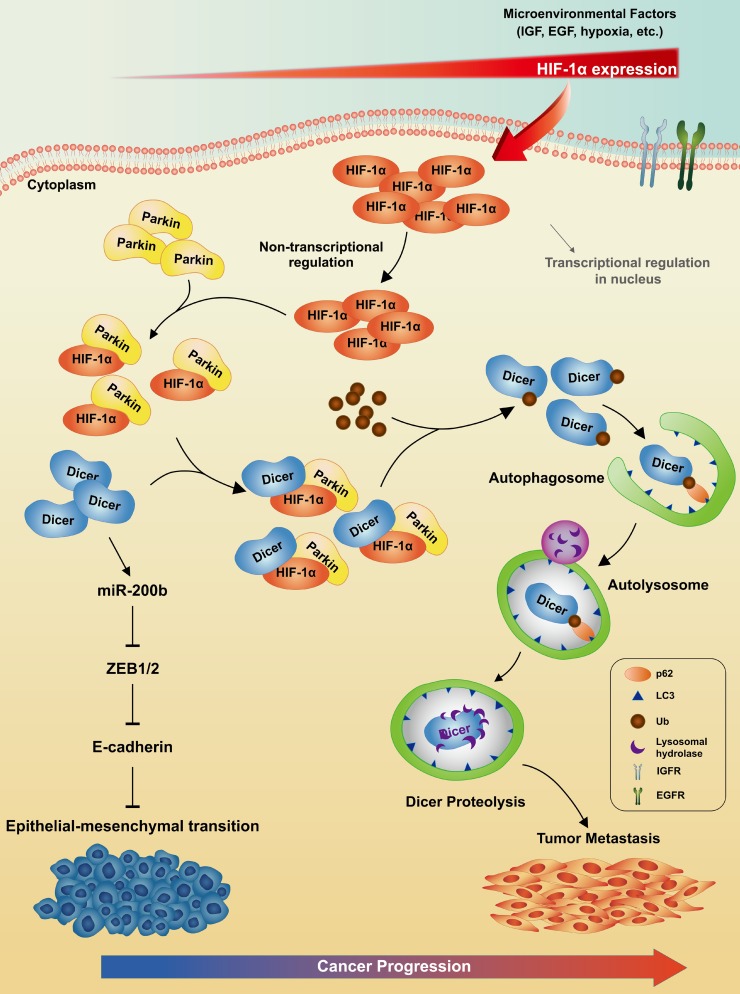
Figure 10. Model for HIF-1α–modulated autophagic proteolysis of Dicer in human cancers.
Microenvironment-induced HIF-1α expression bridges and enhances the binding of Dicer and Parkin. These interactions ubiquitinate Dicer for autophagic-lysosomal proteolysis. Subsequently, down-regulation of Dicer inhibits maturation of miR-200b to promote EMT-induced cancer metastasis.
Ubiquitination is a common posttranslational modification of protein that not only controls protein clearance, but also changes protein localization (53). Polyubiquitination of Dicer has been identified as having a role in the proteasomal degradation pathway (54). Nevertheless, there is a paucity of evidence showing that ubiquitination of Dicer correlates with autophagy degradation and its E3 ligase remains unidentified (55). In the present study, we demonstrate that the Parkin E3 ligase ubiquitinates Dicer, which is subsequently degraded by autophagy-lysosomal proteolysis (Figure 4). This is the first reported evidence, to our knowledge, indicating that Dicer is monoubiquitinated and identifying a potential E3 ligase of Dicer for autophagy. Polyubquitinated Dicer is degraded by the proteasome (54), and monoubquitintated Dicer enters an autophagy-lysosomal pathway, suggesting that different types of ubiquitination may act as switches changing the proteolytic location of Dicer.
The E3 ligase Parkin (PARK2) plays multiple roles in human disease, including Parkinson’s disease and cancer. Homozygous mutations in PARK2 lead to protein misfolding and aggregation, thereby resulting in neuronal integrity damage (56). Furthermore, previous studies have investigated Parkin protein as a potential tumor suppressor that acts by impairing tumorigenesis (57). The heterozygous inactivation of PARK2 is the most common mutation affecting oncogenesis by enhancing cellular growth signals, including those in colon and breast cancer (58, 59). However, several proteins have been identified as substrates of Parkin, including Eps15, Iκκγ, and TRAF2 (60). Parkin-mediated ubiquitination of Eps15 interferes with the binding between Eps15 and EGFR, thereby causing a delay in EGFR endocytosis and activating PI3K/Akt signaling for cell survival (61). Iκκγ and TRAF2 are ubiquitinated by Parkin and confer protection from stress-induced cell death (62). Therefore, the role of Parkin in cancer remains controversial. The results of the present study show that HIF-1α acts as a connector that facilitates the ubiquitination of Dicer through Parkin (Figure 4). In contrast to the tumor-suppressive function of Parkin, HIF-1α controls Parkin and consequently exerts oncogenic effects on cancer metastasis. These mechanisms indicate a potentially novel role of Parkin during cancer progression.
The altered Dicer mRNA was observed only in cells exposed to hypoxia (Supplemental Figure 2I), but not in cells with HIF-1α overexpression/knockdown (Supplemental Figure 2, A and B) or in cells treated with growth factors (Supplemental Figure 2, F and G), suggesting another regulatory mode involving oxygen-sensitive transcriptional regulation, which acts in a HIF-1α–independent manner (Supplemental Figure 2I; ref. 21–23). Reflecting its central role in cellular responses to oxygen-dependent (hypoxia) and oxygen-independent (growth factors and cytokines) stimulations, HIF-1α has been well defined as an oncogenic signaling hub for microenvironmental regulation of cancer progression (1, 5). This study demonstrates a central role of HIF-1α in Dicer regulation under both hypoxic and nonhypoxic conditions. This mode is unexpected on the basis of current knowledge of HIF-1α’s transcriptional activity, and it provides new insights into redefining the biological properties of the well-known transcription factor HIF-1α (Figure 10).
Methods
Cell culture.
The human colorectal cancer cell line (HCT116) was provided by Liang-Yi Hung (Department of Biotechnology and Bioindustry Sciences, NCKU). The human colorectal cancer cell line (Caco-2) and human pancreatic cancer cell lines (Mia-PaCa-2 and PANC1) were provided by Shaw-Jenq Tsai (Department of Physiology, NCKU). Mouse colorectal cancer cell lines (CT-26) were provided by Chih-Peng Chang (Department of Microbiology and Immunology, NCKU). Human breast cancer (MDA-MB-231 and MDA-MB-453), human cervix cancer (HeLa), human lung cancer (A549), and human embryonic kidney (HEK293T) cell lines were obtained from ATCC. Pancreatic cancer cells were grown in RPMI-1640 medium supplemented with 10% FBS (Gibco, Thermo Fisher Scientific). The other cell lines were grown in DMEM/F12 medium supplemented with 10% FBS.
Immunoprecipitation assay.
For regular immunoprecipitation, cells were washed with cold PBS and scraped into NETN lysis buffer (150 mM NaCl, 20 Mm Tris-HCl; pH 8.0, 0.5% NP-40, 1 mM EDTA) containing 1× complete protease inhibitors. For ubiquitination assay, cells were transfected with a FLAG-ubiquitin–expressing plasmid for 24 hours in the presence of NH4Cl (10 mM) and CQ (200 μM). The cells were then washed with cold PBS and scraped into Pierce IP Lysis Buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 5% glycerol, Thermo Fisher Scientific) containing 1× complete protease inhibitors (Roche). After brief sonication, the cell lysates were centrifuged at 14,000 g for 20 minutes at 4°C to remove insoluble cell debris. The cell lysates were subsequently precleaned with protein A/G agarose (for regular immunoprecipitation; Merck) or protein A agarose (for ubiquitination assay; Thermo Scientific) for 1 hour at 4°C. The precleaned cell lysates were mixed with an anti-Dicer antibodies overnight at 4°C and then incubated with agarose beads for 1 hour at 4°C. The immunoprecipitates were assayed by Western blot analysis.
In situ PLA for detection of direct protein-protein interaction.
The in situ PLA was performed according to the manufacturer’s instructions (Sigma-Aldrich). Cancer cells were seeded onto sterile cover slips and grown for 24 hours in a 12-well plate. Tissue arrays containing multiple human normal and tumor tissues were purchased from US Biomax (MNT241). The samples were fixed in 3.7% paraformaldehyde for 20 minutes, permeabilized, and blocked before the addition of the primary antibody for immunofluorescence analysis. After permeabilization, the samples were incubated in the blocking buffer for 1 hour at room temperature in a humidified chamber. The cells were incubated with 2 primary antibodies derived from different species to recognize HIF-1α and Dicer at 4°C overnight. On the following day, the samples were washed in Buffer A 2 times for 5 minutes each and incubated with secondary antibodies conjugated with oligonucleotides for 1 hour at 37°C in a humidified chamber. The ligation reaction was performed at 37°C for 30 minutes. The samples were further incubated with the amplification mixture for 100 minutes at 37°C in a darkened humidified chamber. After washing with 1× Buffer B for 10 minutes, the cells were mounted using mounting medium with DAPI (Vector Laboratories).
In situ PLA for detection of single protein expression.
Protein expression of HIF-1α and Dicer was determined by modified PLA to enhance the specificity. Briefly, human colon cancer tissues were sequentially incubated with 2 specific antibodies recognizing different epitopes of the same protein, following standard manufacturer’s detection procedures. Tissue arrays containing human colon cancer tissues were purchased from Pantomics (COC1021).
Constructs.
pcDNA3.1–HIF-1α was constructed to express HIF-1α with a C-terminal Myc-tag. The following sequences were separately cloned into the pcDNA3.1/Myc-tag vector: HIF-1α with truncated nuclear localization signal (NLS) domain (aa 17–33), HIF-1α with truncated HLH DNA–binding domain (aa 33–71), HIF-1α with truncated Per/Arnt/Sim (PAS) HIF-1β with binding domain (aa 85–298), HIF-1α with truncated oxygen-dependent degradation (ODD) (aa 401–603), HIF-1α with truncated N-terminal activation domain (N-TAD) (aa 531–575), HIF-1α with truncated inhibitory domain (ID) (aa 575–786), and HIF-1α with truncated C-terminal activation domain (C-TAD) (aa 786–826). pcDNA6-ubiquitin was constructed to express ubiquitin with an N-terminal FLAG epitope. pBiFC-VC155–HIF-1α was constructed to express HIF-1α with a C-terminal–fused VC155. pBiFC-VN173-Dicer was constructed to express Dicer with a C-terminal–fused VN173. Mouse WT HIF-1α or HIF-1α ΔHLH was constructed in pLAS2w cDNA expression lentivector. FLAG-tagged Dicer-expressing plasmid was provided by Kohei Miyazono (University of Tokyo, Tokyo, Japan) (63). BiFC-VC155 (no. 22011) and BiFC-VN173 (no. 22010) from Chang-Deng Hu (Purdue University, Lafayette, Indiana, USA) (64), Parkin (no. 45875) and ParkinC431S-expressing (no. 45877) plasmids from Edward Fon (McGill University, Montreal, Canada) (65), truncated Dead (no. 51364), Helicase (no. 541366), DUF283 (no. 51365), and PAZ (no. 51367) Dicer with FLAG-tag plasmids from Narry Kim (Seoul National University, Seoul, South Korea) (66), ZEB1 (no. 35535) from Greg Goodall (University of South Australia, Adelaide, Australia) (41), Lin-41 (no. 11906) from David Bartel (Whitehead Institute for Biomedical Research, Cambridge, Massachusetts, USA) (43), AURKB (no. 29475) from Judy Lieberman (Harvard University, Cambridge, Massachusetts, USA) (44), mRNA 3′ UTR reporters, and miR-200b-200a-429 expression plasmid (no. 35533) from Greg Goodall (41) were obtained from Addgene.
Lentiviral knockdown.
RNA interference was performed using lentiviral shRNAs from RNAi core (Supplemental Table 1; Academia Sinica, Taipei, Taiwan). HEK293T cells were transfected with 5 μg packaging plasmid (pCMVΔR8.91), 0.5 μg envelope plasmid (pMD.G), and 5 μg pLKO.1 shRNA using Lipofectamine 2000 (Invitrogen) for 6 hours. After 24 hours, the supernatants containing viral particles were harvested and filtered through 0.45-μm filters. After infection of lentivirus for 48 hours, cells were selected by puromycin (3 μg/ml). Lentiviral-infected stable clones were maintained in complete medium with puromycin (1.5 μg/ml).
Human subjects.
Human tissue arrays were purchased from US Biomax (MNT241) and Pantomics (COC1021).
Metastasis animal experiments.
Male BALB/c mice (5 to 6 weeks) were purchased from the NCKU laboratory animal center. For intrasplenic injection, 1 × 105 CT-26/Luc cells were suspended in 50 μl PBS and injected into the spleens of mice (n = 7/each experimental group). Mice were intraperitoneally injected with 150 mg/kg of d-luciferin (PerkinElmer), and bioluminescent images were examined by using IVIS noninvasion in vivo imaging system (PerkinElmer). At 18 days after injection, livers and lungs of the mice were isolated to examine the photon intensity of the metastatic signals.
Western blot analysis.
Cells were harvested in RIPA lysis buffer (50 mM Tris-HCl; pH 7.5, 120 mM NaCl, 0.5% NP-40, 200 mM Na3VO4, 1 mM EDTA, 0.5% sodium deoxycholate, 1% SDS) containing 1× protease inhibitor cocktail (Roche) for 10 minutes on ice. An equal amount of protein from the cell lysates was resuspended in gel sample buffer, resolved by SDS-PAGE, and transferred to PVDF membrane (Millipore). After blocking, the membranes were incubated with specific primary antibodies (Supplemental Table 2) overnight at 4°C. After washing with TBST, corresponding secondary antibodies were added at room temperature for 1 hour. The immunoreactivity of the signals was visualized by using an ECL system (PerkinElmer Health Sciences).
MS.
Endogenous HIF-1α was purified through immunoprecipitation with the anti–HIF-1α antibody GTX127309 in HCT116 cells, and samples were subjected to 6% SDS-PAGE. The gels were stained with Coomassie blue. The protein bands corresponding to Dicer (220–240 kDa) and HIF-1α (120 kDa) were excised and subjected to in-gel digestion. The gel was divided into several slices with a scalpel. Gel slices were cut into small pieces, washed with distilled water, and shrunk with acetonitrile. Samples were reduced with DTT and S-alkylated with iodoacetamide, swollen with a solution of 10 ng/μl trypsin (Promega), and incubated overnight at 37°C. After digestion, the supernatants were recovered, and extraction of the remaining peptides from the gel fragments was performed with 5% formic acid, followed by vacuum drying. Peptide desalting was conducted using ZipTip (ZTC18M960, Merck Millipore) and was followed by reverse-phase nano-ultra performance ESI LC-MS/MS, where LS indicates liquid chromatography (SYNAPT G2-Si Mass Spectrometry, Waters) and peptide fragmentation patterning. LC-MS/MS data were obtained using a data-dependent acquisition method and processed by the MASCOT search engine. Data processing for protein identification and quantification was subsequently performed using SWISS-PROT databases.
Confocal microscopy analysis.
Cells were washed 3 times with cold PBS, fixed in 4% formaldehyde for 1 hour, and then permeated with 0.5% Triton X-100 for 15 minutes and incubated with 5% BSA for 1 hour. Afterwards, cells were incubated with primary antibodies (diluted at 1:200) overnight at 4°C. On the following day, cells were washed by PBS and further incubated with the appropriate secondary antibody (diluted at 1:200) tagged with rhodamine or FITC (Millipore) at room temperature for 1 hour. For BiFC, cells were transfected with VC155–HIF-1α and VN173-fused Dicer. Excitation of Venus was detected at 515 nm (67). Nuclei were stained with DAPI together with mounting medium (Vector Laboratories). All images were taken by using a Carl Zeiss LSM780 laser-scanning confocal microscope from the Instrument Development Center, NCKU.
Boyden chamber migration assay.
Boyden chamber cell migration assays were performed using uncoated Transwells (Corning). The Transwells were inserted into 24-well plates containing DMEM/F12 supplemented with 10% FBS. HCT116 cells were transfected with HIF-1α or ΔHLH HIF-1α–expressing plasmid for 24 hours. The following day, cells were transfected with miR-200b mimics (MDbio) for 24 hours. After trypsinization, cells were seeded into Transwells for 48 hours, and subsequently, the nonmigrated cells were removed with a cotton swab and the migrated cells were fixed with 4% formaldehyde for 1 hour and further stained with crystal violet. The migrated cells on the lower surface of the membrane were observed under an optical microscope. The number of migrated cells was quantified by blind counting from at least 3 fields per chamber under a 20× objective.
Isolation of RNA and reverse-transcription real-time PCR.
Total RNA was isolated by using TRIzol (Invitrogen), and cDNA was synthesized by miRNA-specific reverse-transcription primer (for miRNA) or random hexamers (for pri-mRNA and mRNA). The cDNA of miRNA and pri-mRNA were amplified by using Universal PCR Master Mix with TaqMan probe (Applied Biosystems). The PCR cycling parameters were set according to the manufacturer’s protocol. The amplificational replication signal was detected by Applied Biosystems Step One Real-Time PCR System. Values representing the average of 3 independent experiments were normalized to the endogenous control gene. The cDNA of mRNA was amplified by specific primers (Supplemental Table 3) with Taq polymerase. The PCR cycling parameters were as follows: 95°C for 3 minutes and 35 cycles of 95°C for 40 seconds, 60°C for 40 seconds, and 75°C for 30 seconds.
Autophagosome isolation.
Extraction was performed as previously reported (68). Briefly, HCT116 cells were transfected with HIF-1α for 48 hours. The cells were suspended in a hypotonic solution (10% sucrose mixed with 0.5 ml 1 M HEPES/0.1 M EDTA) and homogenized with 20 strokes of a syringe (23-gauge). The resultant homogenate was diluted with homogenization buffer (0.25 M sucrose, 10 mM HEPES, 1 mM EDTA, pH 7.3) containing 1.5 mM glycyl-l-phenylalanine 2-naphthylamide (GPN) and 1% DMSO. After incubation for 7 minutes at 37°C to destroy the lysosomes, the homogenate was cooled to 4°C.
Statistics.
All data were analyzed using Prism 6 (GraphPad), and the results are presented as mean ± SD using at least 3 independent experiments. Unpaired 2-tailed Student’s t test was used to assess 2 independent groups. One-way ANOVA was used to test multigroup comparisons with Tukey’s post hoc test. The groups with 2 independent variables were tested by 2-way ANOVA (Figure 3B, Figure 6F, and Supplemental Figure 3B). P < 0.05 was considered statistically significant.
Study approval.
All protocols of animal studies using BALB/c mice and operation of the IVIS spectrum followed the principles of 3Rs and were approved by the IACUC of NCKU. Tissue arrays and frozen colon cancer tissues obtained from NCKU Hospital were used according to IRB protocols approved by the NCKU Hospital. Experiments using human subjects were approved by the NCKU Hospital IRB. All human subjects gave informed consent.
Author contributions
HHL and PSC conceived and designed all experiments. CFC and HHL conducted MS analysis. HHL identified the functional domains. JNL conducted PLA analysis and pri-miRNA detection. HHL, JNL, MYW, JWK, and HYH collected and assayed tumor samples. HHL, JNL, and YJL performed all of the other experiments in this study. HIS designed and established FLAG-tagged Dicer-expressing plasmid. HLS, MCH, CMC, and PSC discussed the data and wrote this paper.
Supplementary Material
Acknowledgments
We thank Liang-Yi Hung, Chih-Peng Chang, and Hsiao-Sheng Liu for their technical support. We thank Kohei Miyazono for providing FLAG-tagged Dicer-expressing plasmid. We thank Kalle Gehring, V. Narry Kim, Gregory J. Goodall, Christopher B. Burge, Ashish Lal, and Judy Lieberman for sharing their plasmids with Addgene. We are grateful to Sheng-Hsiang Lin for providing statistical consulting services from the Biostatistics Consulting Center, NCKU Hospital. We are grateful for the support from the Human Biobank, Research Center of Clinical Medicine, and the Cancer Data Bank of NCKU Hospital. We thank the National RNAi Core Facility at Academia Sinica in Taiwan for providing shRNA reagents and related services. Mass spectrometry analyses were performed by Research Facility for Sharing at NHRI, Taiwan (Core Facilities for Proteomics and Chemistry). We thank the staff of the Biomedical Resource Core at the First Core Labs, National Taiwan University College of Medicine, for technical assistance. This study was supported by the Ministry of Science and Technology, Taiwan (MOST 103-2628-B-006-003-MY3, MOST 105-2320-B-006-050, MOST 106-2320-B-006-038, MOST 106-2320-B-006-021).
Version 1. 12/18/2017
Electronic publication
Version 2. 02/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(2):625–643.https://doi.org/10.1172/JCI89212.
Contributor Information
Hui-Huang Lai, Email: like_jerry@hotmail.com.
Jie-Ning Li, Email: jessy61621@hotmail.com.
Ming-Yang Wang, Email: suryang1971@hotmail.com.
Hsin-Yi Huang, Email: hyhuang4010@ntu.edu.tw.
Hui-Lung Sun, Email: huilung@uchicago.edu.
Yu-Jhen Lyu, Email: sallyliu96@hotmail.com.
Jui-Wen Kang, Email: kang1594@gmail.com.
Ching-Feng Chiu, Email: chodino1106@gmail.com.
Mien-Chie Hung, Email: mhung@mdanderson.org.
Pai-Sheng Chen, Email: bio.benson@gmail.com.
References
- 1.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 2.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129(3):465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12(12):5447–5454. doi: 10.1128/MCB.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270(3):1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 5.LaGory EL, Giaccia AJ. The ever-expanding role of HIF in tumour and stromal biology. Nat Cell Biol. 2016;18(4):356–365. doi: 10.1038/ncb3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005;2005(306):re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- 7.Beck I, Ramirez S, Weinmann R, Caro J. Enhancer element at the 3’-flanking region controls transcriptional response to hypoxia in the human erythropoietin gene. J Biol Chem. 1991;266(24):15563–15566. [PubMed] [Google Scholar]
- 8.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 10.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 12.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khoshnaw SM, et al. Loss of Dicer expression is associated with breast cancer progression and recurrence. Breast Cancer Res Treat. 2012;135(2):403–413. doi: 10.1007/s10549-012-2169-3. [DOI] [PubMed] [Google Scholar]
- 14.Faggad A, et al. Down-regulation of the microRNA processing enzyme Dicer is a prognostic factor in human colorectal cancer. Histopathology. 2012;61(4):552–561. doi: 10.1111/j.1365-2559.2011.04110.x. [DOI] [PubMed] [Google Scholar]
- 15.Martello G, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141(7):1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 16.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 17.Kumar MS, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23(23):2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villanueva T. Metastasis: It’s nicer with DICER. Nat Rev Cancer. 2010;10(8):530–531. doi: 10.1038/nrc2910. [DOI] [PubMed] [Google Scholar]
- 19.Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014;6(1):211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2(1):38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 21.van den Beucken T, et al. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun. 2014;5:5203. doi: 10.1038/ncomms6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandara V, Michael MZ, Gleadle JM. Hypoxia represses microRNA biogenesis proteins in breast cancer cells. BMC Cancer. 2014;14:533. doi: 10.1186/1471-2407-14-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho JJ, et al. Functional importance of Dicer protein in the adaptive cellular response to hypoxia. J Biol Chem. 2012;287(34):29003–29020. doi: 10.1074/jbc.M112.373365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12(9):836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 25.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 26.Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33(4):505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 27.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10(11):1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 28.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399(6733):271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 29.Chen D, Li M, Luo J, Gu W. Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function. J Biol Chem. 2003;278(16):13595–13598. doi: 10.1074/jbc.C200694200. [DOI] [PubMed] [Google Scholar]
- 30.Arndt V, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20(2):143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 31.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 32.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirsilä M, Koivunen P, Günzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem. 2003;278(33):30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- 34.Epstein AC, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107(1):43–54. doi: 10.1016/S0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 35.Pongratz I, Antonsson C, Whitelaw ML, Poellinger L. Role of the PAS domain in regulation of dimerization and DNA binding specificity of the dioxin receptor. Mol Cell Biol. 1998;18(7):4079–4088. doi: 10.1128/MCB.18.7.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hogenesch JB, et al. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J Biol Chem. 1997;272(13):8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 37.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 38.Cifuentes D, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328(5986):1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang X, Le QT, Giaccia AJ. MiR-210--micromanager of the hypoxia pathway. Trends Mol Med. 2010;16(5):230–237. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulshreshtha R, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27(5):1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 42.Yu F, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 43.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 44.Lal A, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’UTR microRNA recognition elements. Mol Cell. 2009;35(5):610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Francia G, Cruz-Munoz W, Man S, Xu P, Kerbel RS. Mouse models of advanced spontaneous metastasis for experimental therapeutics. Nat Rev Cancer. 2011;11(2):135–141. doi: 10.1038/nrc3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim C, Cauchy F, Azoulay D, Farges O, Ronot M, Pocard M. Tumour progression and liver regeneration--insights from animal models. Nat Rev Gastroenterol Hepatol. 2013;10(8):452–462. doi: 10.1038/nrgastro.2013.55. [DOI] [PubMed] [Google Scholar]
- 47.Urosevic J, et al. Colon cancer cells colonize the lung from established liver metastases through p38 MAPK signalling and PTHLH. Nat Cell Biol. 2014;16(7):685–694. doi: 10.1038/ncb2977. [DOI] [PubMed] [Google Scholar]
- 48.Goddard ET, Fischer J, Schedin P. A portal vein injection model to study liver metastasis of breast cancer. J Vis Exp. 2016;(118):54903. doi: 10.3791/54903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee WY, Hong HK, Ham SK, Kim CI, Cho YB. Comparison of colorectal cancer in differentially established liver metastasis models. Anticancer Res. 2014;34(7):3321–3328. [PubMed] [Google Scholar]
- 50.Li Y, et al. Suppression of cancer relapse and metastasis by inhibiting cancer stemness. Proc Natl Acad Sci U S A. 2015;112(6):1839–1844. doi: 10.1073/pnas.1424171112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Obacz J, Pastorekova S, Vojtesek B, Hrstka R. Cross-talk between HIF and p53 as mediators of molecular responses to physiological and genotoxic stresses. Mol Cancer. 2013;12(1):93. doi: 10.1186/1476-4598-12-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460(7254):529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 53.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315(5809):201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Z, et al. Valproic acid causes proteasomal degradation of DICER and influences miRNA expression. PLoS ONE. 2013;8(12):e82895. doi: 10.1371/journal.pone.0082895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O. Corrigendum: Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2015;17(8):1088. doi: 10.1038/ncb3208. [DOI] [PubMed] [Google Scholar]
- 56.Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 57.Devine MJ, Plun-Favreau H, Wood NW. Parkinson’s disease and cancer: two wars, one front. Nat Rev Cancer. 2011;11(11):812–823. doi: 10.1038/nrc3150. [DOI] [PubMed] [Google Scholar]
- 58.Cesari R, et al. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc Natl Acad Sci U S A. 2003;100(10):5956–5961. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Veeriah S, et al. Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet. 2010;42(1):77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sandebring A, Cedazo-Mínguez A. Parkin- An E3 ubiquitin ligase with multiple substrates. J Alzheimers Dis Parkinsonism. 2012:S10:002. https://doi.org/10.4172/2161-0460.S10-002.
- 61.Fallon L, et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat Cell Biol. 2006;8(8):834–842. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- 62.Henn IH, et al. Parkin mediates neuroprotection through activation of IkappaB kinase/nuclear factor-kappaB signaling. J Neurosci. 2007;27(8):1868–1878. doi: 10.1523/JNEUROSCI.5537-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki HI, et al. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell. 2011;44(3):424–436. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 64.Shyu YJ, Liu H, Deng X, Hu CD. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques. 2006;40(1):61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 65.Trempe JF, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340(6139):1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 66.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25(3):522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shyu YJ, Hiatt SM, Duren HM, Ellis RE, Kerppola TK, Hu CD. Visualization of protein interactions in living Caenorhabditis elegans using bimolecular fluorescence complementation analysis. Nat Protoc. 2008;3(4):588–596. doi: 10.1038/nprot.2008.16. [DOI] [PubMed] [Google Scholar]
- 68.Seglen PO, Brinchmann MF. Purification of autophagosomes from rat hepatocytes. Autophagy. 2010;6(4):542–547. doi: 10.4161/auto.6.4.11272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.