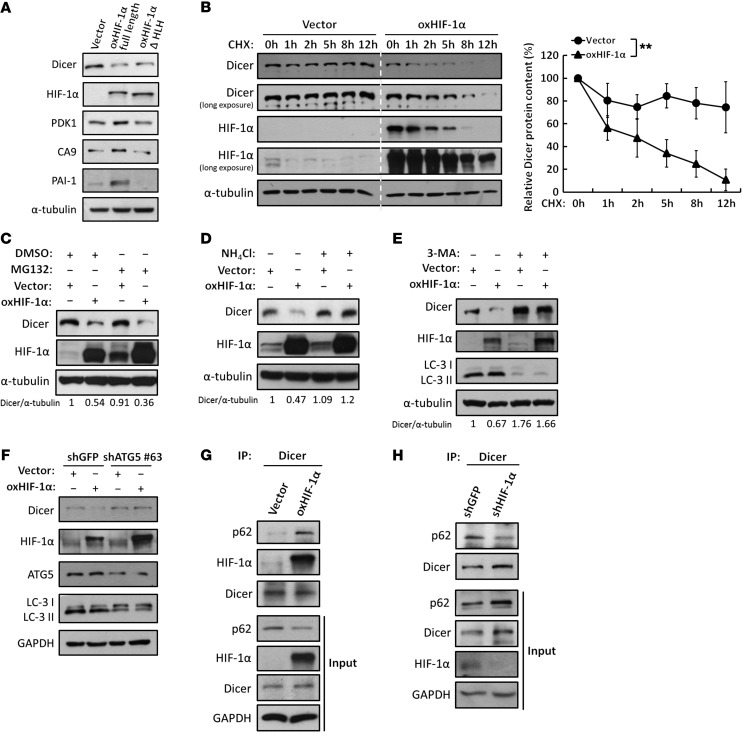
Figure 3. HIF-1α enhances proteolysis of Dicer through an autophagy-lysosomal pathway.
(A) Effects of the nontranscriptional activity of HIF-1α on Dicer. WT HIF-1α or HLH-truncated HIF-1α was overexpressed in HCT116 cells for Western blot analysis to determine the expression of Dicer, PDK1, CA9, and PAI-1. The immunoblots presented were derived from replicate samples run on parallel gels. (B) HIF-1α enhances the protein degradation of Dicer. Cells were treated with CHX at the indicated times to block de novo protein synthesis. The expression of Dicer was determined by Western blot analysis and was further quantified by ImageJ (NIH). The relative levels of Dicer protein remaining at each time point were normalized to α-tubulin (right). Data are presented as mean ± SD, with at least n = 3 per group. **P < 0.01, 2-way ANOVA. All of the cell lysates were run on the same gel. (C–E) Effects of the proteolytic pathways on the HIF-1α–induced downregulation of Dicer. Cells were treated with MG132 to inhibit proteasomal degradation (C), ammonium chloride (NH4Cl) to inhibit lysosomal degradation (D), or 3-MA to inhibit autophagic degradation (E). The immunoblots presented were derived from replicate samples run on parallel gels. The ratios of Dicer/α-tubulin protein levels were quantified by using ImageJ. (F) Similar experiments were performed in cells in which ATG5 was genetically knocked down to inhibit autophagy. LC3 was detected for autophagic activation. (G and H) The associations between Dicer and the autophagy receptor p62 were analyzed in the presence of NH4Cl and CQ to block autophagy-lysosomal degradation in HIF-1α–overexpressing (G) or HIF-1α knockdown (H) HCT116 cells. The immunoblots presented were derived from replicate samples run on parallel gels (H).