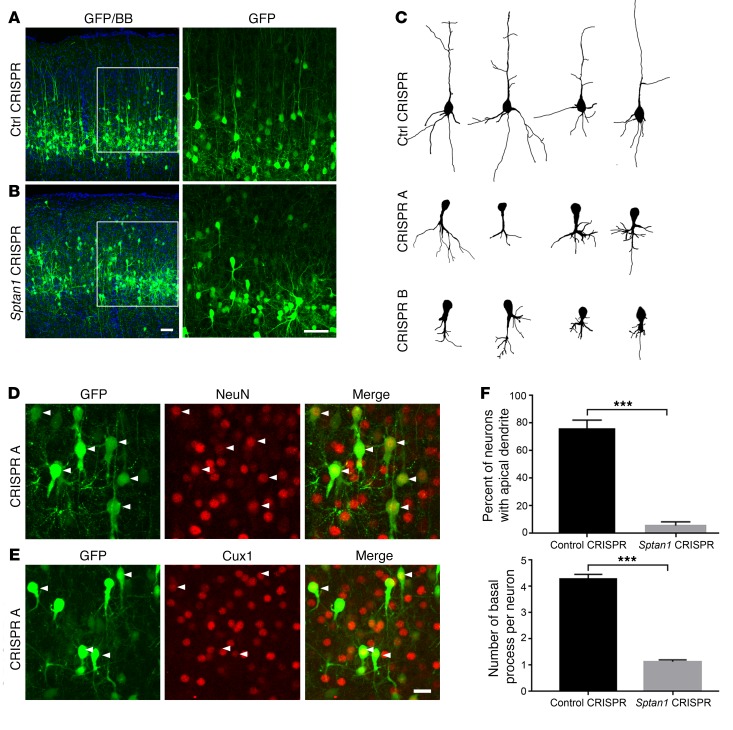
Figure 3. Altered neuronal morphology after Sptan1 deletion.
(A) Confocal images of P21 brain sections after control CRISPR transfection at E14–15 show normal-appearing GFP-labeled cells in cortical layer II/III (left). A higher-magnification image of the boxed region (right) reveals typical pyramidal neuron morphology with a single apical dendrite and multiple basal dendrites. (B) Neurons transfected with Sptan1 CRISPR A reach the appropriate cortical layers but appear disorganized, lack apical dendrites, and have abnormal basal dendritic architecture that typically consists of a single short, thickened but exuberant process. (C) Reconstructed neurons transfected with control CRISPR or Sptan1 CRISPR A or B. Note that neurons transfected in vivo with Sptan1 CRISPR A or B are morphologically similar, indicating the specificity of the gene editing. (D and E) Although Sptan1 deletion severely alters neuronal morphology, the GFP+ cells coexpress the neuronal marker NeuN (red in D) and the superficial cortical layer marker Cux1 (red in E), suggesting preserved neuronal differentiation. Scale bars: 25 μm in B (for A and B), 10 μm in E (for D and E). (F) Quantification of percentages of GFP-labeled cells with apical dendrites and numbers of basal dendrites per GFP+ cell in neurons transfected with control plasmid or Sptan1 CRISPR A. A total of 39 neurons from 3 brains with Sptan1 CRISPR A transfection and 43 neurons from 3 brains from the CRISPR control group were quantified. Percentage data are transformed to arcsine value for statistical analysis. One-tailed t test: ***P < 0.001.