Figure 4. Deletion of YY1 Binding Sites Causes Loss of Enhancer-Promoter Interactions.
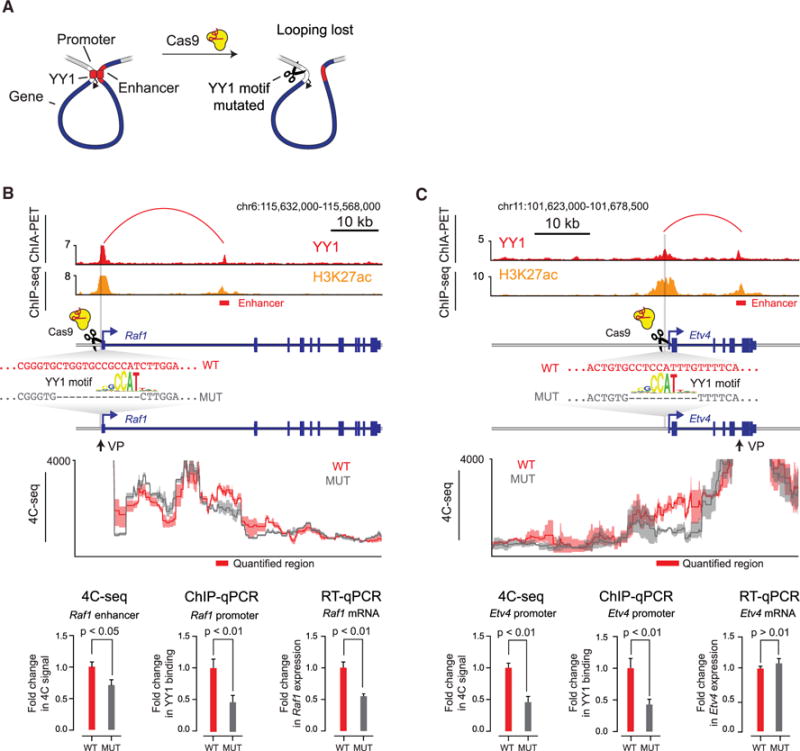
(A) Model depicting CRISPR/Cas9-mediated deletion of a YY1 binding motif in the regulatory region of a gene.
(B and C) CRISPR/Cas9-mediated deletion of YY1 binding motifs in the regulatory regions of two genes, Raf1 (B) and Etv4 (C), was performed, and the effects on YY1 occupancy, enhancer-promoter looping, and mRNA levels were measured. The positions of the targeted YY1 binding motifs, the genotype of the wild-type and mutant lines, and the 4C sequencing (4C-seq) viewpoint are indicated. The mean 4C-seq signal is represented as a line (individual replicates are shown in Figure S4), and the shaded area represents the 95% confidence interval. Three biological replicates were assayed for 4C-seq and ChIP-qPCR experiments, and six biological replicates were assayed for RT-qPCR experiments. Error bars represent the SD. All p values were determined using the Student’s t test.
See also Figure S4. See STAR Methods for detailed description of genomics analyses. Datasets used in this figure are listed in Table S4.
