Summary
Clustered regularly interspaced short palindromic repeats‐associated protein 9 (CRISPR‐Cas9) is a revolutionary technology that enables efficient genomic modification in many organisms. Currently, the wide use of Streptococcus pyogenes Cas9 (SpCas9) primarily recognizes sites harbouring a canonical NGG protospacer adjacent motif (PAM). The newly developed VQR (D1135V/R1335Q/T1337R) variant of Cas9 has been shown to cleave sites containing NGA PAM in rice, which greatly expanded the range of genome editing. However, the low editing efficiency of the VQR variant remains, which limits its wide application in genome editing. In this study, by modifying the single guide RNA (sgRNA) structure and strong endogenous promoters, we significantly increased the editing efficiency of the VQR variant. The modified CRISPR‐Cas9‐VQR system provides a robust toolbox for multiplex genome editing at sites containing noncanonical NGA PAM.
Keywords: CRISPR‐Cas9, VQR, genome editing, efficiency, rice
Introduction
The clustered regularly interspaced short palindromic repeats‐associated protein 9 (CRISPR‐Cas9) system with a single guide RNA (sgRNA) has exhibited powerful capabilities for genome editing in eukaryotes and is widely used for investigating the function of genes or rapidly obtaining new alleles (Cong et al., 2013; Hsu et al., 2014; Mali et al., 2013; Shan et al., 2013). Editing efficiency is affected by many factors, including the low GC contents of the guide sequence, secondary structures of target sequences (Ma et al., 2015b; Wang et al., 2014; Zhang et al., 2014) and continuous A or T on the guide sequence (Dang et al., 2015). Thus, although the CRISPR‐Cas9 provides a robust system for genome editing, it remains inefficient or hardly applicable in many genomic sites. To increase the editing efficiency in animal cells or embryos, pre‐assembled Cas9 protein–gRNA ribonucleoproteins (RNPs) are frequently used. Recently, it was found that nonhomologous single‐stranded DNA could stimulate a disruption frequency, and that the sgRNA structure could be optimized to increase the efficiency of the CRISPR‐Cas9 system in mammalian cells (Dang et al., 2015; Kim et al., 2014; Lin et al., 2014; Richardson et al., 2016a,b). In addition, RNA‐guided TevCas9 dual nuclease was developed, which could increase the efficiency of the CRISPR‐Cas9 system by creating a deletion of small fragments (Wolfs et al., 2016). In regard to plants, the main strategy employed to increase the editing efficiency of the CRISPR‐Cas9 system is to use strong promoters to drive high expression of Cas9 and to avoid usage of a low‐scored guide sequence (Ma et al., 2015b; Shan et al., 2013; Zhang et al., 2014).
Currently, the robust and widely used Streptococcus pyogenes Cas9 (SpCas9) requires the sites containing NGG protospacer adjacent motifs (PAMs) (Hsu et al., 2014). To expand the range of CRISPR‐Cas9 genome editing, many orthologs or variants of Cas9 proteins were screened out, such as S. thermophilus CRISPR3 Cas9 for NGGNG PAMs (Horvath et al., 2008), S. thermophilus CRISPR1 Cas9 for NNAGAAW PAMs (Deveau et al., 2008), Neisseria meningitidis Cas9 for NNNNGATT PAMs (Zhang et al., 2013), and VQR variants of SpCas9 for NGA PAMs and VRER variants for NGCG PAMs (Kleinstiver et al., 2015). In plants, the VQR and VRER variants of Cas9 for genome editing have been developed, which greatly extended the range of genome editing (Hu et al., 2016). However, the efficiency of these variants remains low compared with that of wild‐type (WT) Cas9, which limits its wide application for genome editing.
In this study, we aimed to increase the editing efficiency of the CRISPR‐Cas9‐VQR system. By modifying the sgRNA structure and expressing VQR variants with strong endogenous promoters, we dramatically increased the editing efficiency of VQR variants in rice. Thus, the newly modified CRISPR‐Cas9‐VQR system is particularly suitable for efficient genome editing in NGA PAMs.
Results
We first modified the sgRNA structure to that in mammalian cells (Dang et al., 2015) and tested whether the modification is able to increase the editing efficiency of CRISPR/Cas9 in plants. The previously widely used gRNAs contain continuous sequences of Ts, which are the pausing signals for RNA polymerase III and efficiently reduce transcription. We replaced the fourth T in the sequence of Ts with C to eliminate the pausing signal. In addition, five pairs of bases were added into the distal duplex of the sgRNA to enhance its stability or binding to the Cas9 protein (Dang et al., 2015) (Figure 1a). We screened the rice genome and selected two sites containing NGG PAMs in the MONOCULM 3 (MOC3) and GRAIN WIDTH ON CHROMOSOME 2 (GW2) genes for genome editing (Lu et al., 2015; Song et al., 2007). Both guide sequences were calculated to be low‐scored sites (Ma et al., 2016; Xu et al., 2015) (Table S1). Furthermore, the guide sequence of GW2 also contains low GC (31.58%) and a continuous sequence close to the PAM, both of which would affect the editing efficiency. The guide sequences were cloned into a modified and unmodified sgRNA, respectively. The two guide sequences using the same sgRNAs were then assembled into one vector (Figure 1b). The constructed vectors were then transferred into rice by an Agrobacterium‐mediated method (Hiei et al., 1994). A total of 88 and 92 independent transgenic lines using unmodified and modified sgRNAs were obtained, respectively. By DNA sequencing, we found that all target sites harboured mutations with different mutation rate (Tables S2 and S3). The mutation rate at MOC3 (from 65.91% to 64.13%) remained stable, whereas that at GW2 increased from 44.32% to 56.52%. As for the frequency of mutants (biallelic or chimeric mutants), the mutant proportion at the MOC3 site almost doubled, increasing from 23.86% to 45.65%. However, the mutant proportion at the GW2 site increased more than fivefold, from 6.82% to 34.78% (Figure 1c, d and Table S3). Furthermore, the proportion of moc3 gw2 double mutants dramatically increased by approximately 13‐fold (from 2.27% to 29.35%) (Figure 1e and Table S4). These results indicate that the modified sgRNAs are functional in promoting the efficiency of Cas9 at sites containing NGG PAMs in plants.
Figure 1.
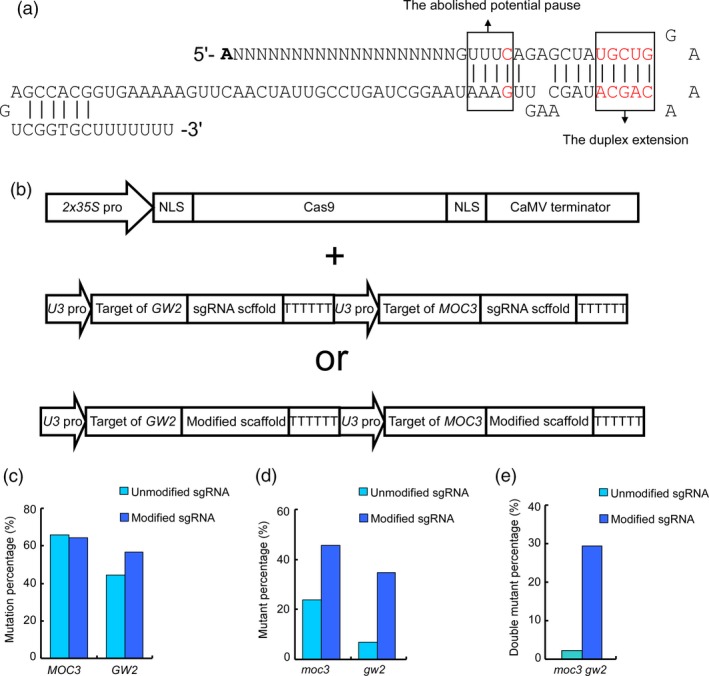
Modified single guide RNAs (sgRNAs) increased the efficiency of the clustered regularly interspaced short palindromic repeats‐associated protein 9 (CRISPR‐Cas9) system. (a) Schematic representation of modified sgRNAs. The replaced or introduced nucleotides are highlighted in red. These mutations abolish a potential transcription pause site and add a duplex extension for sgRNA stability. (b) The architecture of vectors in the CRISPR‐Cas9 system. The Cas9 protein attached to the nuclear localization signal (NLS) is driven by the 2x35S promoter. Both unmodified and modified sgRNAs are expressed by the U3 promoter. (c) The mutation rates at MOC3 and GW2. The modified sgRNA and unmodified sgRNA are represented in dark blue and light blue, respectively. (d) The proportion of mutants (biallelic or chimeric mutants) of moc3 and gw2. By modifying sgRNAs, the proportion of mutants increased approximately twofold or fivefold at MOC3 and GW2, respectively. (e) The proportion of moc3 gw2 double mutants. Modified sgRNAs dramatically increased (approximately 13‐fold) the proportion of double mutants.
We next investigated whether the modified sgRNAs promote the editing efficiency at NGA PAM sites in the CRISPR‐Cas9‐VQR system. The guide sequence, which has been shown to simultaneously edit three genomic sites harbouring NGA PAMs (Hu et al., 2016), was selected. These sites were distinguished by a different base (A, T and G) behind the NGA PAMs. We ligated the guide sequence into the modified sgRNA, which was then assembled into the vector expressing VQR variants (Figure 2a). The resulting vector was subsequently used for genetic transformation, and a total of 43 transgenic lines were obtained. By sequencing all three target sites, we analysed the editing efficiencies and compared the data with those obtained from unmodified sgRNA. By using the modified sgRNAs, the mutation rate at target‐A increased marginally from 4.08% to 4.65%, whereas at target‐T, the rate dramatically quintupled, increasing from 2.04% to 9.30%, and at target‐G, the rate nearly doubled, increasing from 18.37% to 30.23% (Figure 2b, Tables S5 and S6). We then analysed the plants harbouring double mutations and found that the frequency of double mutations increased sharply from 2.04% to 13.95% using the modified sgRNA (Figure 2b and Table S7). These results indicate that the modified sgRNAs are effective in improving the efficiency of the CRISPR‐Cas9‐VQR system.
Figure 2.
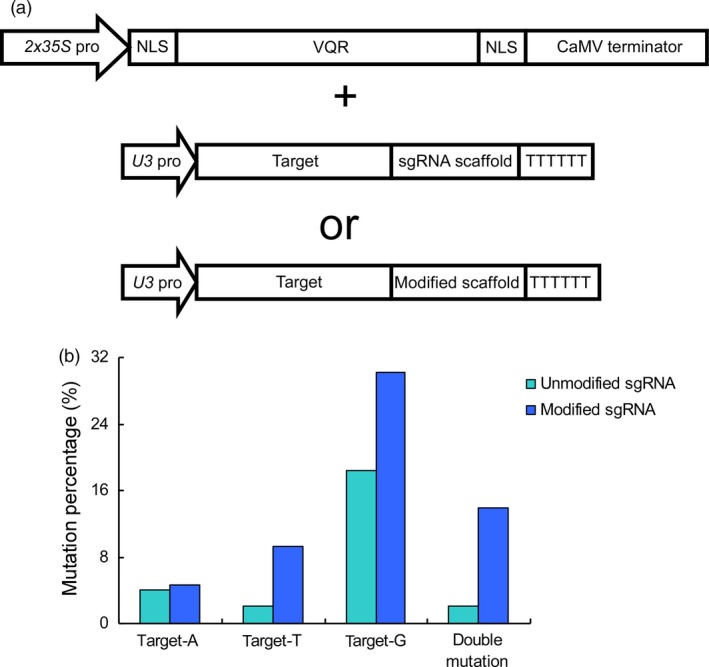
Efficiency of the CRISPR‐Cas9‐VQR system is promoted by modified sgRNAs. (a) The architecture of vectors in the CRISPR‐Cas9‐VQR system. The VQR variants and sgRNAs are expressed by the 2x35S and U3 promoters, respectively. The binary vectors are constructed using one of the two sgRNAs. (b) The proportions of single mutations and double mutations. A, T and G, respectively, indicate the base following the NGA PAM. The unmodified sgRNA and modified sgRNA are represented in light blue and dark blue, respectively.
In our previous studies, we utilized a 2x35S promoter to drive the expression of VQR protein (Hu et al., 2016). To further increase the efficiency of the CRISPR‐Cas9‐VQR system, we replaced the 2x35S promoter with two endogenous promoters, namely rice UBIQUITIN1 (UBI1) and ACTIN1 (ACT1), respectively (McElroy et al., 1990; Wang et al., 2000) (Figures S1 and S2). For comparison, the same guide sequence described above was used further for genome editing. After vector construction (Figure 3a) and genetic transformation, 36 transgenic lines using the UBI1 or ACT1 promoter were obtained, respectively (Tables S8 and S9). We sequenced the target sites of all transgenic plants and compared the editing efficiency with that obtained by using unmodified sgRNAs and the 2x35S promoter. When the UBI1 promoter and the modified sgRNA were co‐applied, the mutation rate at target‐A increased from 4.08% to 19.44%, whereas that at target‐T increased from 2.04% to 11.11% and that at target‐G increased from 18.37% to 38.89% (Figure 3b and Table S10). Furthermore, eight lines were detected to possess biallelic mutations at target‐G sites, which were not found using the original system (Tables S8 and S10). Moreover, the proportion of lines containing double and triple mutations increased from 2.04% to 19.44% (Figure 3c and Table S6).
Figure 3.
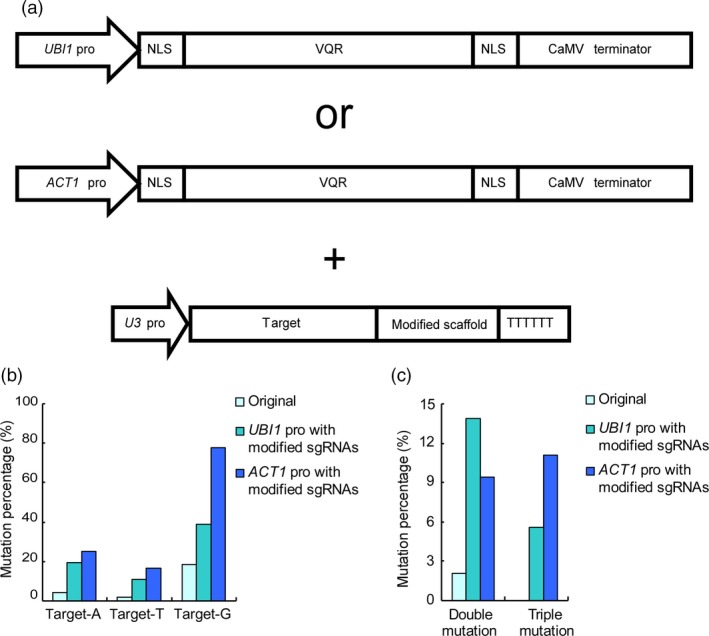
Efficiency of the CRISPR‐Cas9‐VQR system is further increased by the UBI1 and ACT1 promoters. (a) The architecture of vectors with different promoters. The promoters of VQR variants are replaced by UBI1 or ACT1. The modified sgRNAs are expressed with U3 promoters. (b) The mutation rate of the CRISPR‐Cas9‐VQR system using different promoters. The efficiency of the CRISPR‐Cas9‐VQR system is greatly increased by using strong promoters and modified sgRNAs. The system with the UBI1 promoter increases the mutation rate by an average of approximately fourfold, whereas that with ACT1 increases the mutation rate by an average of approximately sixfold. (c) The proportion of double and triple mutations using different systems. The proportions of double or triple mutations are dramatically increased. The triple mutations are undetected in the original system.
When the ACT1 promoter and modified sgRNA were used, the editing efficiency at target‐A increased approximately sixfold from 4.08% to 25.00%, that at target‐T increased approximately eightfold from 2.04% to 16.67%, and that at target‐G increased approximately fourfold from 18.37% to 77.78% (Figure 3b and Table S10). In addition, more than half (52.78%) of transgenic plants were identified as biallelic mutants (Figure 3 and Table S10). Moreover, the proportion of transgenic lines harbouring double and triple mutations increased by approximately 15‐fold from 2.04% to 30.56% (Figure 3c and Table S6). The results imply that both UBI1 and ACT1 promoters can significantly promote the editing efficiency of the CRISPR‐Cas9‐VQR system. In particular, the ACT1 promoter is the most efficient among the three promoters in elevating the editing efficiency of the CRISPR‐Cas9‐VQR system (Figure 3b and Table S10).
A previous study of the CRISPR‐Cas9 system indicated that increased efficiency would raise the risk of off‐targeting effects (Singh et al., 2016). To investigate whether similar patterns exist in the CRISPR‐Cas9‐VQR system, we screened the rice genome and selected four sites for analysis. All four sites contained only one mismatch with the target sequence (Table 1). The mismatch was located at the 19‐, 17‐, 15‐ or 6‐base pair (bp) position from NAG PAMs, respectively. By sequencing all these sites in the modified lines, we found that off‐targeting events occurred at VQR‐off‐1, VQR‐off‐2 and VQR‐off‐3 sites, but not at VQR‐off‐4 sites. In addition, the off‐targeting events showed an obvious preference (VQR‐off‐1 > VQR‐off‐2 > VQR‐off‐3) in each line (Table 1, Figure S3 and Table S11). We also compared the off‐targeting events between lines showing different editing efficiency and found that with the increased editing efficiency, the frequency of off‐targeting events increased accordingly (Figure S3).
Table 1.
The off‐target events in the CRISPR‐Cas9‐VQR system
| Name of potential off‐target site | Location | Sequence | Guide RNA | Promoters | No. of plants sequenced | No. of modified plants | Mutation rate (%) |
|---|---|---|---|---|---|---|---|
| VQR‐off‐1 | 11:18102183‐18102203 | AGCGGCGGCGGCGGCGTCAAGA | Unmodified | 2x35S | 11 | 9 | 81.82 |
| Modified | 2x35S | 13 | 13 | 100 | |||
| UBI1 | 16 | 16 | 100 | ||||
| ACT1 | 28 | 28 | 100 | ||||
| VQR‐off‐2 | 10:15997834:15998455 | GGTGGCGGCGGCGGCGTCAGGA | Unmodified | 2x35S | 11 | 6 | 54.55 |
| Modified | 2x35S | 13 | 12 | 92.31 | |||
| UBI1 | 16 | 16 | 100 | ||||
| ACT1 | 28 | 28 | 100 | ||||
| VQR‐off‐3 | 5:25504329:25504950 | GGCGTCGGCGGCGGCGTCATGA | Unmodified | 2x35S | 11 | 1 | 9.09 |
| Modified | 2x35S | 13 | 0 | 0 | |||
| UBI1 | 16 | 2 | 12.50 | ||||
| ACT1 | 28 | 1 | 3.57 | ||||
| VQR‐off‐4 | 6:13414921:13415542 | GGCGGCGGCGGCGCCGTCAAGA | Unmodified | 2x35S | 11 | 0 | 0 |
| Modified | 2x35S | 13 | 0 | 0 | |||
| UBI1 | 16 | 0 | 0 | ||||
| ACT1 | 28 | 0 | 0 |
The protospacer adjacent motif (PAM) and the mismatch nucleotides are highlighted in blue and red, respectively.
Discussion
In the present study, by the modification of sgRNAs and replacement of rice strong promoters, we revealed that the CRISPR‐Cas9‐VQR system is efficient in genome editing at sites containing NGA PAMs.
The genome editing using the CRISPR‐Cas9 system requires two components: a Cas9 nuclease to cut the target sequence and a sgRNA that binds to the target sequence. The robustness of the system is highly correlated with the level of expression of both the components (Ma et al., 2015b). Previous studies in human cells showed that the mutation of continuous Ts and the extension of the duplex of sgRNA could result in increased sgRNA production, which significantly improves the editing efficiency of the system (Dang et al., 2015). We adopted a similar strategy to modify the sgRNA structure and found that it can also improve the efficiency of genome editing in both the CRISPR‐Cas9 and CRISPR‐Cas9‐VQR systems. In addition to modified sgRNA, the rice strong endogenous promoters could further enhance the editing efficiency of the CRISPR‐Cas9‐VQR system, particularly with the ACT1 promoter that could increase the average editing frequency nearly sixfold compared with the original system. Moreover, the frequency of edited plants containing double or triple mutations increased approximately 15‐fold. Previous studies revealed that the editing efficiency of the VQR variant at NGAA or NGAT PAMs is lower than that at NGAG PAM (Kleinstiver et al., 2015). Our study found that the increases in editing efficiency at the sites containing NGAA or NGAT PAMs are more significant than those harbouring NGAG sites, suggesting that the inefficient PAMs might be more sensitive to the expression levels of the VQR variant and sgRNA.
Various studies using the CRISPR‐Cas9 system showed that a high editing efficiency is frequently accompanied by a high frequency of off‐targeting events (Fu et al., 2013; Hsu et al., 2013; Wang et al., 2015b; Zhang et al., 2014). We also found that the closer the mismatch to the PAMs, the lower the off‐targeting frequency. Our results demonstrated that a similar pattern exists in the CRISPR‐Cas9‐VQR system. Therefore, the off‐targeting should be taken into account when the efficient CRISPR‐Cas9‐VQR system is used for genome editing.
Collectively, we modified the CRISPR‐Cas9‐VQR system and significantly increased the efficiency of genome editing. The work provided a more powerful system for genome editing at the sites harbouring noncanonical NGA PAMs in plants.
Experimental procedures
Modification of sgRNA
The modification used is based on SK‐gRNA, which assembled the expression cassette of sgRNAs described in our previous study (Wang et al., 2015a). The SK‐gRNA digested with two restriction enzymes (SalI and kpnI) and the big fragment was purified as the vector. The expression cassette of modified sgRNAs was divided into two small fragments for polymerase chain reaction (PCR) with KOD FX DNA Polymerase (TOYOBO, Japan) and connected into the vector. Modified‐1‐F and Modified‐1‐R were designed for one of two small fragments, whereas Modified‐2‐F and Modified‐2‐R were designed for the remaining fragments (Table S12). The template used was SK‐gRNA, and the thermocycler was set for one cycle of 94 °C for 2 min, 32 cycles of 98 °C for 10 s, 58 °C for 30 s, 68 °C for 30 s and one cycle of 68 °C for 5 min, and maintained at 4 °C. The recombination of the vector and two small fragments was conducted with the ClonExpress MultiS One Step Cloning Kit (Vazyme, Nanjing, China).
Construction of expressing plasmids
The sgRNA‐Cas9 expressing plasmids were constructed using the isocaudamer ligation method (Wang et al., 2015a). The sgRNAs of MOC3 (digested with KpnI/SalI) and GW2 (digested with XhoI/BglII) were assembled in one pC1300‐Cas9 binary vector (digested with KpnI/BamHI). The sgRNA for VQR variants (digested with KpnI/BglII) was assembled into pC1300‐2x35S/UBI1 pro‐VQR binary vector (digested with KpnI/BamHI), respectively. In addition, another sgRNA for VQR variants (digested with KpnI/NheI) was assembled into pC1300‐ACT1 pro‐VQR binary vector (digested with KpnI/XbaI).
The generation of transgenic rice
The rice (Oryza sativa L. ssp. japonica) variety Nipponbare was used as the host plant in the present study. The strain EHA105 was used for generation of transgenic rice by the Agrobacterium‐mediated method, which was conducted as previously described (Hiei et al., 1994).
Replacement of promoters
The ACT1 promoter was amplified from pcambia2300Actin vector using ACT1‐F/ACT1‐R primers, whereas the UBI1 promoter was amplified from the rice genome using UBI1‐F/UBI1‐R primers (Table S12). The promoters were recombined into pC1300‐VQR (digested with KpnI/NcoI) by Gibson Assembly.
PCR amplification of target regions and sequencing
The primers used for PCR amplification are listed in Table S12 with KOD FX DNA Polymerase (TOYOBO, Japan). The product of PCR was sequenced by the Sanger method, and the multiple peaks were decoded by the Degenerate Sequence Decoding (DSD) method (Ma et al., 2015a).
Author Contributions
K.W. and J.L. designed the studies. X.H. and X.M. performed the experiments. Q.L. conducted the bioinformatic analyses. K.W., J.L., X.H. and X.M. wrote the manuscript.
Competing Financial Interests
The authors declare no conflict of interests.
Supporting information
Figure S1 The sequence of UBI1 promoter.
Figure S2 The sequence of ACT1 promoter.
Figure S3 Off‐target effects of different systems in modified plants. Four potential off‐target sites with one mismatch are detected in all modified plants.
Table S1 The primers and oligos used to construct the sgRNA.
Table S2 The results of sequence modification using CRISPR‐ Cas9 system.
Table S3 Comparison of mutations at MOC3 and GW2 sites using unmodified and modified sgRNAs.
Table S4 Comparison of double mutations at MOC3 and GW2 sites using unmodified and modified sgRNAs.
Table S5 The results of sequence modification by VQR using modified sgRNA and 2x35S promoter.
Table S6 Comparison of the mutations in CRISPR‐Cas9‐VQR system using unmodified and modified sgRNAs.
Table S7 Comparison of double and triple mutations in CRISPR‐Cas9‐VQR system.
Table S8 The results of sequence modification by VQR using modified sgRNA and UBI1 promoter.
Table S9 The results of sequence modification by VQR using modified sgRNA and ACT1 promoter.
Table S10 Comparison of the mutations using different promoters in CRISPR‐Cas9‐VQR system.
Table S11 The results of off‐target with VQR.
Table S12 The primers used in the study.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (3140101312, 91635310 and 31271681) and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences.
Contributor Information
Jiayang Li, Email: jyli@genetics.ac.cn.
Kejian Wang, Email: wangkejian@caas.cn.
References
- Cong, L. , Ran, F.A. , Cox, D. , Lin, S. , Barretto, R. , Habib, N. , Hsu, P.D. et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang, Y. , Jia, G. , Choi, J. , Ma, H. , Anaya, E. , Ye, C. , Shankar, P. et al (2015) Optimizing sgRNA structure to improve CRISPR‐Cas9 knockout efficiency. Genome Biol. 16, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau, H. , Barrangou, R. , Garneau, J.E. , Labonte, J. , Fremaux, C. , Boyaval, P. , Romero, D.A. et al (2008) Phage response to CRISPR‐encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. , Foden, J.A. , Khayter, C. , Maeder, M.L. , Reyon, D. , Joung, J.K. and Sander, J.D. (2013) High‐frequency off‐target mutagenesis induced by CRISPR‐Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei, Y. , Ohta, S. , Komari, T. and Kumashiro, T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Horvath, P. , Romero, D.A. , Coute‐Monvoisin, A.C. , Richards, M. , Deveau, H. , Moineau, S. , Boyaval, P. et al (2008) Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190, 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, P.D. , Scott, D.A. , Weinstein, J.A. , Ran, F.A. , Konermann, S. , Agarwala, V. , Li, Y. et al (2013) DNA targeting specificity of RNA‐guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, P.D. , Lander, E.S. and Zhang, F. (2014) Development and applications of CRISPR‐Cas9 for genome engineering. Cell, 157, 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X. , Wang, C. , Fu, Y. , Liu, Q. , Jiao, X. and Wang, K. (2016) Expanding the range of CRISPR/Cas9 genome editing in rice. Mol Plant, 9, 943–945. [DOI] [PubMed] [Google Scholar]
- Kim, S. , Kim, D. , Cho, S.W. , Kim, J. and Kim, J.S. (2014) Highly efficient RNA‐guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver, B.P. , Prew, M.S. , Tsai, S.Q. , Topkar, V.V. , Nguyen, N.T. , Zheng, Z. , Gonzales, A.P. et al (2015) Engineered CRISPR‐Cas9 nucleases with altered PAM specificities. Nature, 523, 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. , Staahl, B.T. , Alla, R.K. and Doudna, J.A. (2014) Enhanced homology‐directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife, 3, e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Z. , Shao, G. , Xiong, J. , Jiao, Y. , Wang, J. , Liu, G. , Meng, X. et al (2015) MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J Genet Genom. 42, 71–78. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Chen, L. , Zhu, Q. , Chen, Y. and Liu, Y.G. (2015a) Rapid decoding of sequence‐specific nuclease‐induced heterozygous and biallelic mutations by direct sequencing of PCR products. Mol Plant, 8, 1285–1287. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Zhang, Q. , Zhu, Q. , Liu, W. , Chen, Y. , Qiu, R. , Wang, B. et al (2015b) A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Mol Plant, 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Ma, J. , Koster, J. , Qin, Q. , Hu, S. , Li, W. , Chen, C. , Cao, Q. et al (2016) CRISPR‐DO for genome‐wide CRISPR design and optimization. Bioinformatics, 32, 3336–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P. , Yang, L. , Esvelt, K.M. , Aach, J. , Guell, M. , DiCarlo, J.E. , Norville, J.E. et al (2013) RNA‐guided human genome engineering via Cas9. Science, 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy, D. , Zhang, W. , Cao, J. and Wu, R. (1990) Isolation of an efficient actin promoter for use in rice transformation. Plant Cell, 2, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, C.D. , Ray, G.J. , Bray, N.L. and Corn, J.E. (2016a) Non‐homologous DNA increases gene disruption efficiency by altering DNA repair outcomes. Nat. Commun. 7, 12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, C.D. , Ray, G.J. , DeWitt, M.A. , Curie, G.L. and Corn, J.E. (2016b) Enhancing homology‐directed genome editing by catalytically active and inactive CRISPR‐Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34, 339–344. [DOI] [PubMed] [Google Scholar]
- Shan, Q. , Wang, Y. , Li, J. , Zhang, Y. , Chen, K. , Liang, Z. , Zhang, K. et al (2013) Targeted genome modification of crop plants using a CRISPR‐Cas system. Nat. Biotechnol. 31, 686–688. [DOI] [PubMed] [Google Scholar]
- Singh, D. , Sternberg, S.H. , Fei, J. , Doudna, J.A. and Ha, T. (2016) Real‐time observation of DNA recognition and rejection by the RNA‐guided endonuclease Cas9. Nat. Commun. 7, 12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X.J. , Huang, W. , Shi, M. , Zhu, M.Z. and Lin, H.X. (2007) A QTL for rice grain width and weight encodes a previously unknown RING‐type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Jiang, J. and Oard, J.H. (2000) Structure, expression and promoter activity of two polyubiquitin genes from rice (Oryza sativa L.). Plant Sci. 156, 201–211. [DOI] [PubMed] [Google Scholar]
- Wang, T. , Wei, J.J. , Sabatini, D.M. and Lander, E.S. (2014) Genetic screens in human cells using the CRISPR‐Cas9 system. Science, 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Shen, L. , Fu, Y. , Yan, C. and Wang, K. (2015a) A simple CRISPR/Cas9 system for multiplex genome editing in rice. J Genet Genom. 42, 703–706. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Wang, Y. , Wu, X. , Wang, J. , Wang, Y. , Qiu, Z. , Chang, T. et al (2015b) Unbiased detection of off‐target cleavage by CRISPR‐Cas9 and TALENs using integrase‐defective lentiviral vectors. Nat. Biotechnol. 33, 175–178. [DOI] [PubMed] [Google Scholar]
- Wolfs, J.M. , Hamilton, T.A. , Lant, J.T. , Laforet, M. , Zhang, J. , Salemi, L.M. , Gloor, G.B. et al (2016) Biasing genome‐editing events toward precise length deletions with an RNA‐guided TevCas9 dual nuclease. Proc. Natl. Acad. Sci. USA, 113, 14988–14993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Xiao, T. , Chen, C.H. , Li, W. , Meyer, C.A. , Wu, Q. , Wu, D. et al (2015) Sequence determinants of improved CRISPR sgRNA design. Genome Res. 25, 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Heidrich, N. , Ampattu, B.J. , Gunderson, C.W. , Seifert, H.S. , Schoen, C. , Vogel, J. et al (2013) Processing‐independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol. Cell, 50, 488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zhang, J. , Wei, P. , Zhang, B. , Gou, F. , Feng, Z. , Mao, Y. et al (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The sequence of UBI1 promoter.
Figure S2 The sequence of ACT1 promoter.
Figure S3 Off‐target effects of different systems in modified plants. Four potential off‐target sites with one mismatch are detected in all modified plants.
Table S1 The primers and oligos used to construct the sgRNA.
Table S2 The results of sequence modification using CRISPR‐ Cas9 system.
Table S3 Comparison of mutations at MOC3 and GW2 sites using unmodified and modified sgRNAs.
Table S4 Comparison of double mutations at MOC3 and GW2 sites using unmodified and modified sgRNAs.
Table S5 The results of sequence modification by VQR using modified sgRNA and 2x35S promoter.
Table S6 Comparison of the mutations in CRISPR‐Cas9‐VQR system using unmodified and modified sgRNAs.
Table S7 Comparison of double and triple mutations in CRISPR‐Cas9‐VQR system.
Table S8 The results of sequence modification by VQR using modified sgRNA and UBI1 promoter.
Table S9 The results of sequence modification by VQR using modified sgRNA and ACT1 promoter.
Table S10 Comparison of the mutations using different promoters in CRISPR‐Cas9‐VQR system.
Table S11 The results of off‐target with VQR.
Table S12 The primers used in the study.


