Figure 1. Conditional Excision of Cdc73 Induces Differentiation and Alters Epigenetic Landscape in MA9 Leukemic Cells.
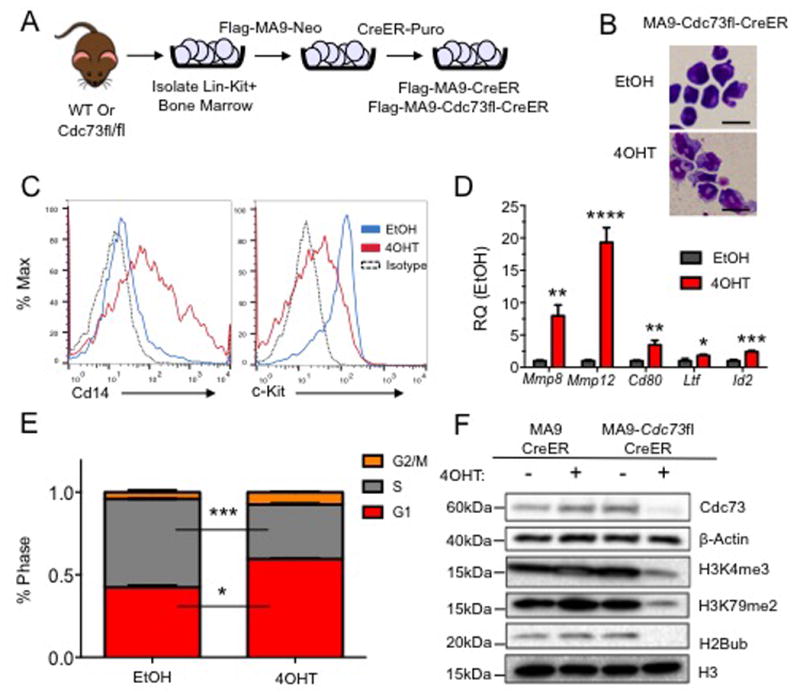
A) Scheme showing generation of Flag-tagged MA9 leukemic Cdc73 excision cells driven by inducible CreER for phenotypic analysis. B) MA9-Cdc73fl-CreER cells were treated with either EtOH or 7.5nM 4OHT for 48 hours, cytospun onto slides and stained with the Hema3 staining kit. Representative images at 100× show morphology changes. Scale bar = 20μM C) Excision of Cdc73 increases level of differentiation as measured by c-Kit, Cd14 cell surface markers on MA9 leukemic cells by flow cytometry. D) Cdc73 excision increases differentiation gene signature expression including Mmp8, Mmp12, Cd80, Ltf and Id2. All samples were scaled to β-actin and normalized to EtOH control expression. * = p-value < 0.05, ** = p-value < 0.01, *** = p-value < 0.001, **** = p-value < 0.0001. Student t-test. E) Cdc73 excision results in a G1 cell cycle block. MA9 cells were treated as in (B) and cell cycle analysis performed using ModFit software. * = p-value < 0.05, *** = p-value < 0.001 Student t-test. F) Excision of Cdc73 alters global epigenetic landscape. Cells were treated as in (B), and ∼2×106 cells were harvested after 72 hours and subjected to histone extraction and probed with the indicated antibodies. H3 and β-Actin are used as loading controls. All experiments were performed in biological duplicate and technical duplicate for phenotypic analysis or technical triplicate for qPCR.
