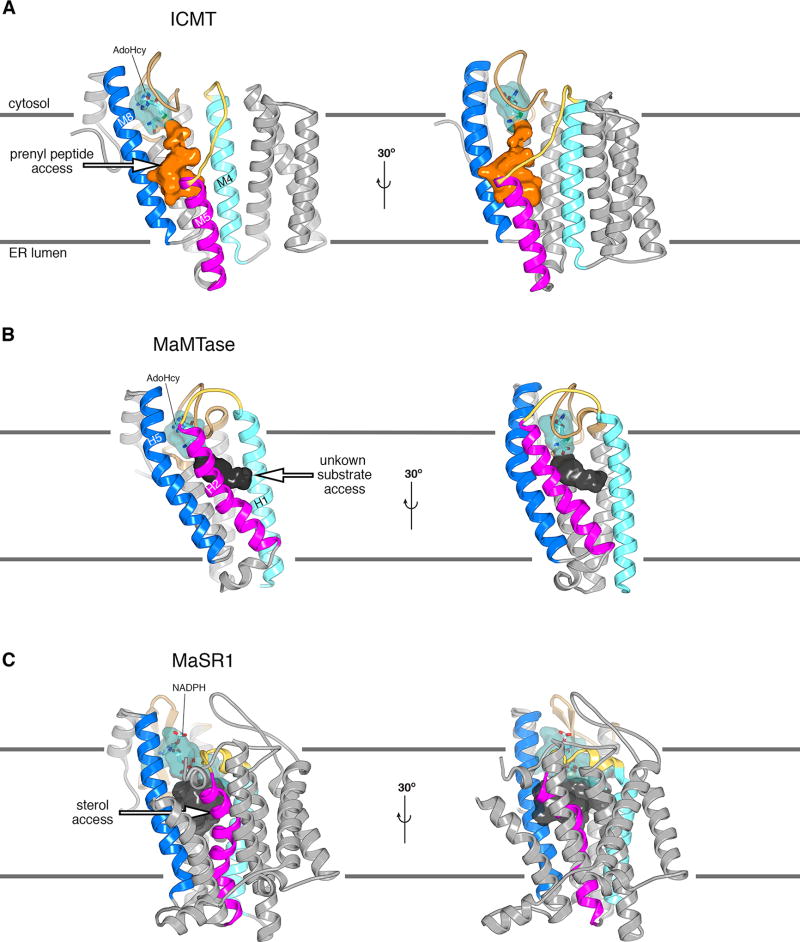
Extended Data Figure 8. Comparison of the active sites of ICMT, MaMTase and MaSR1.
a, Two orientations of the overall structure of ICMT are shown with the active site depicted as a molecular surface. The portion that binds AdoHcy is coloured cyan and is partially transparent to reveal a stick representation of the cofactor. The cavity that would be exposed to the membrane and forms the presumed binding site of the prenylcysteine moiety of the substrate is coloured orange. An arrow marks a pathway by which the prenyl group of the substrate could reach the active site via the membrane. Horizontal lines denote the approximate boundaries of the ER membrane. b, Overall structure of MaMTase (a prokaryotic integral membrane methyltransferase previously termed “Ma-ICMT” in 16, PDB ID: 4A2N) shown in two orientations, which were obtained by superposing with ICMT. A cavity, denoting the active site, is drawn as a molecular surface. The portion in which AdoHcy binds is coloured cyan; the remainder is dark grey and likely represents the binding site of its biological substrates, which are unknown. Structural elements are coloured as for ICMT in a. The cavity is exposed to the membrane on the opposite side of the enzyme relative to ICMT. Instead of having a crevice between the magenta- and dark blue-coloured helices like ICMT (H2 and H5 of MaMTase, which roughly correspond to M5 and M8 of ICMT, respectively), MaMTase has a crevice between H1 (light blue) and H2 (magenta), suggesting that its substrates access it from the “right” (arrow) rather than from the “left”. In ICMT, this route is blocked by the M4–M5 linker. The dimensions of the cavity in MaMTase and its exposure to the membrane suggest that the substrates of MaMTase are hydrophobic molecules with smaller dimensions than a farnesyl or geranylgeranyl prenyl group. c, Overall structure of the prokaryotic integral membrane sterol reductase MaSR1 (PDB ID: 4QUV)18. The orientation is based on superposition of the cofactor binding domain with ICMT, with corresponding colouring. The carbon atoms of a bound NADPH cofactor are coloured cyan. A crevice between the magenta- and dark blue-coloured helices may serve as access for lipophilic sterol substrates (arrow)18.