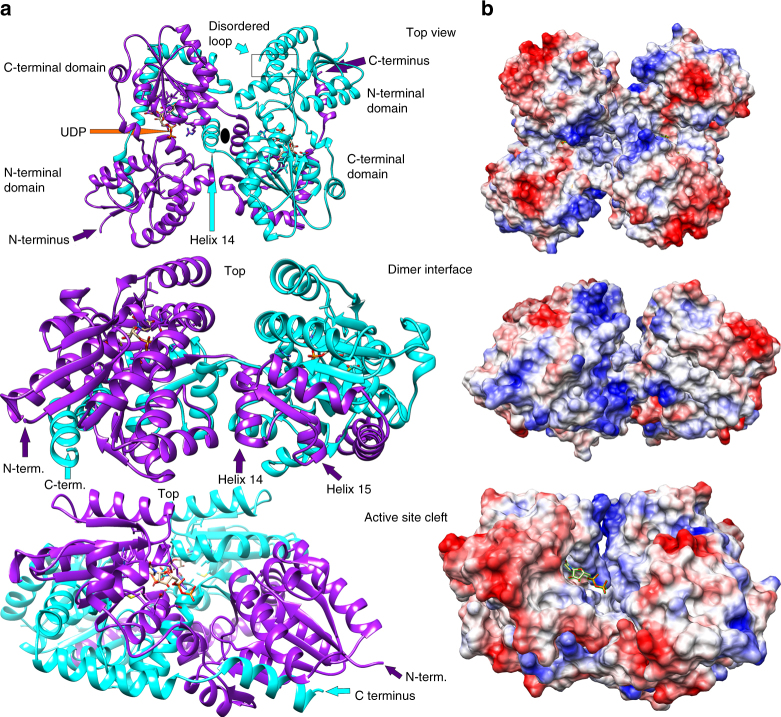
Fig. 2.
Overall structure of LpxB. a The overall structure of LpxB7S shows that LpxB forms a dimer in which the C-terminus of one subunit completes the fold of the opposite subunit. The polypeptides are exchanged at helix 14. Each domain forms a Rossmann-like fold with a parallel β-sheet surrounded by α-helices. UDP is bound to the C-terminal domain showing the position of the UDP-binding pocket where the sugar donor substrate, UDP-DAG, binds. Lipid X likely binds to the N-terminal domain which includes the predicted catalytic base D9810. LpxB appears to be in an open conformation. Catalysis may involve partial closing of the cleft between the domains by a hinge-like movement of the N-terminal domain similar to conformational changes observed for MurG38, PimA50, and other GT-B enzymes40. b Columbic surface rendering (blue for positive and red for negative) corresponding to the adjacent ribbons in a. The surface near the dimer interface is highly basic and is likely involved in membrane association