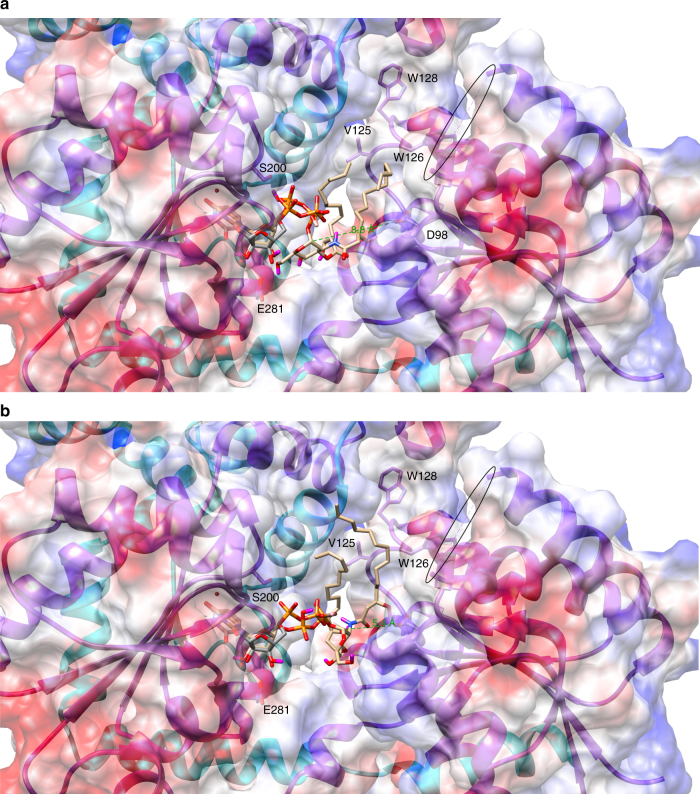
Fig. 6.
Molecular docking model of UDP-DAG binding. UDP-DAG (tan carbons) was docked into the LpxB active site cleft with AutoDock Vina56. The software correctly identified the nucleotide-binding site overlaying reasonably well with the position of UDP (gray carbons) observed in the crystal structure. The lipid tails were predicted to extend toward a hydrophobic groove lined by V125, W126, and W128. The break in the chain corresponding to the disordered loop is circled. a The top ranked model places the anomeric carbon 8.8 Å from the predicted catalytic base, D98. b A lower ranked model (seven of nine) places the anomeric carbon 5.4 Å from D98, which would require less movement of the protein in order to correctly orient D98 and the 6′-hydroxyl of lipid X for proton transfer and nucleophilic attack on the anomeric carbon