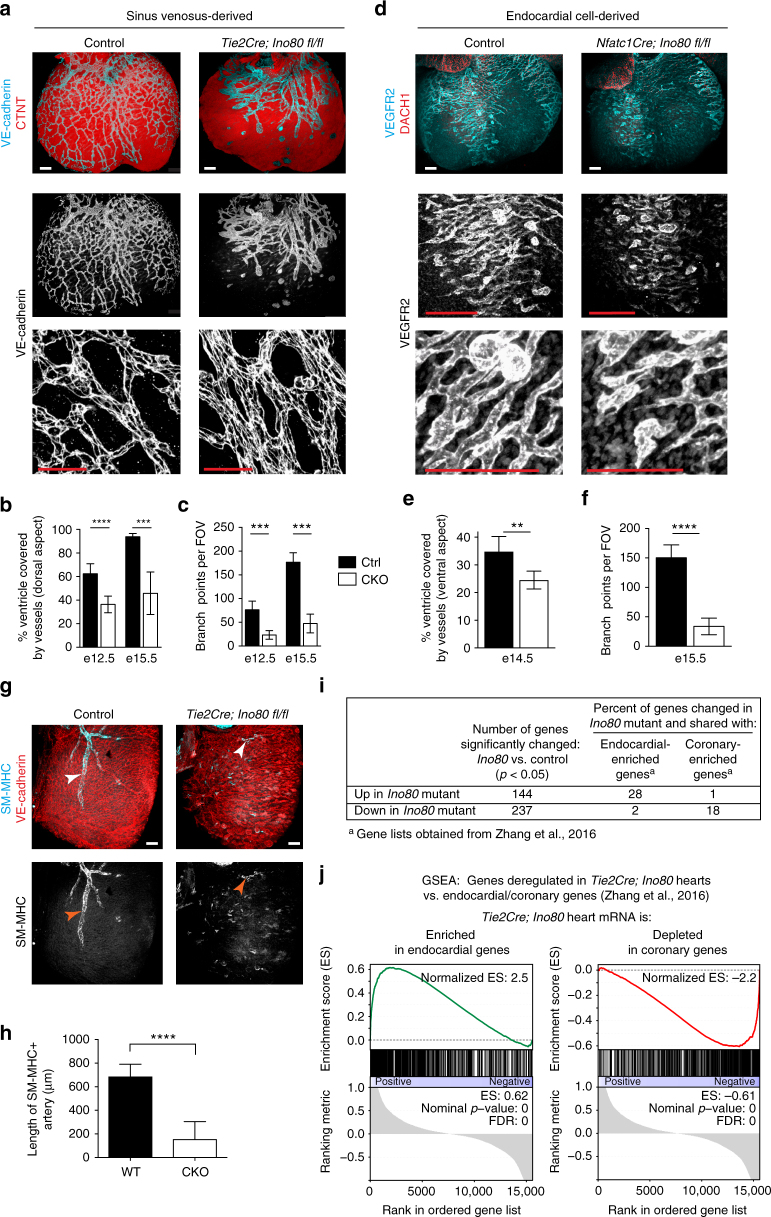
Fig. 4.
In vivo angiogenesis from both the sinus venosus and endocardial cells is defective with Ino80 deletion. a–f Whole-mount confocal images (a, d) and quantification (b, c, e, f) of coronary vessel growth in indicated Ino80-deficient hearts. a, b VE-cadherin immunofluorescence (a) and quantification of heart coverage (b) revealed that vessel growth on the dorsal side of the heart where the sinus venosus sprouts is significantly stunted. Images are representative of the following number of replicates: control, n = 4 hearts; mutant, n = 7 hearts at e15.5. Error bars in graphs are sd. (control, n = 8 hearts; mutant n = 6 hearts at e12.5, control, n = 4 hearts; mutant, n = 7 hearts at e15.5). ****P < 0.0001, evaluated by Student’s t-test. Scale bars: 100 μm. c Vessel branching is also reduced in whole hearts. Error bars in graphs are sd. (control, n = 5 hearts; mutant, n = 6 hearts at e12.5, control, n = 4 hearts; mutant, n = 7 hearts at e15.5). ***P < 0.001, evaluated by Student’s t-test. d, e VEGFR2 immunofluorescence (d) and vessel coverage (e) shows a similar significant decrease on the ventral side of the heart where endocardial cell-derived coronary migration occurs. Images are representative of the following number of replicates: control, n = 6 hearts; mutant, n = 5 hearts at e15.5. Error bars in graphs are standard deviation. (control, n = 6 hearts; mutant, n = 5 hearts at e15.5). **P < 0.01, evaluated by Student’s t-test. Scale bars: 100 um. f Endocardial-derived vessel branching is also stunted in whole hearts. Error bars in graphs are standard deviation. (control, n = 4 hearts; mutant, n = 6 hearts at e15.5). ****P < 0.0001, evaluated by Student’s t-test. Scale bars: 100 μm. g, h Confocal images of the right lateral side of e15.5 hearts (g) and quantification (h) showed a dramatic decrease in SM-MHC+ arterial smooth muscle (arrowheads) in mutant hearts. Error bars in graphs are standard deviation. Images are representative of the following number of replicates: control, n = 5 hearts; mutant, n = 6 hearts. Scale bars: 100 μm. i, j Comparison of genes changed in e13.5 Tie2Cre;Ino80 fl/fl hearts with those reported to be specifically expressed in either endocardial or coronary endothelial cells. Ino80 mutants are enriched in endocardial genes and depleted of coronary genes