Fig. 1.
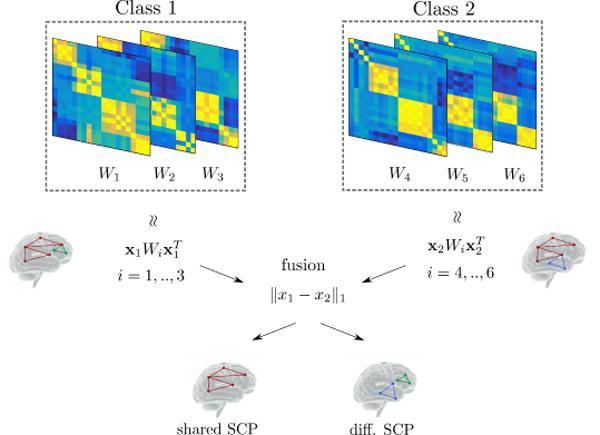
Simplified illustration of the proposed method in the case of two classes. Using correlation matrices from each class, we extract sets of co-activated sub-networks (Sparse Connectivity Patterns, or SCP) represented by the components of vectors xk, k = 1, 2. In this simple case, a single SCP (i.e. column vector) is estimated for each class. These SCP’s are fused across classes during estimation depending on the weight parameter λ2 in Eq. 3. This allows us to extract SCP components that are shared across classes (red sub-network) with an increased analysis power, since the whole dataset is used. This in turns facilitates further identification of differential/class-specific SCP (green and blue sub-networks).
