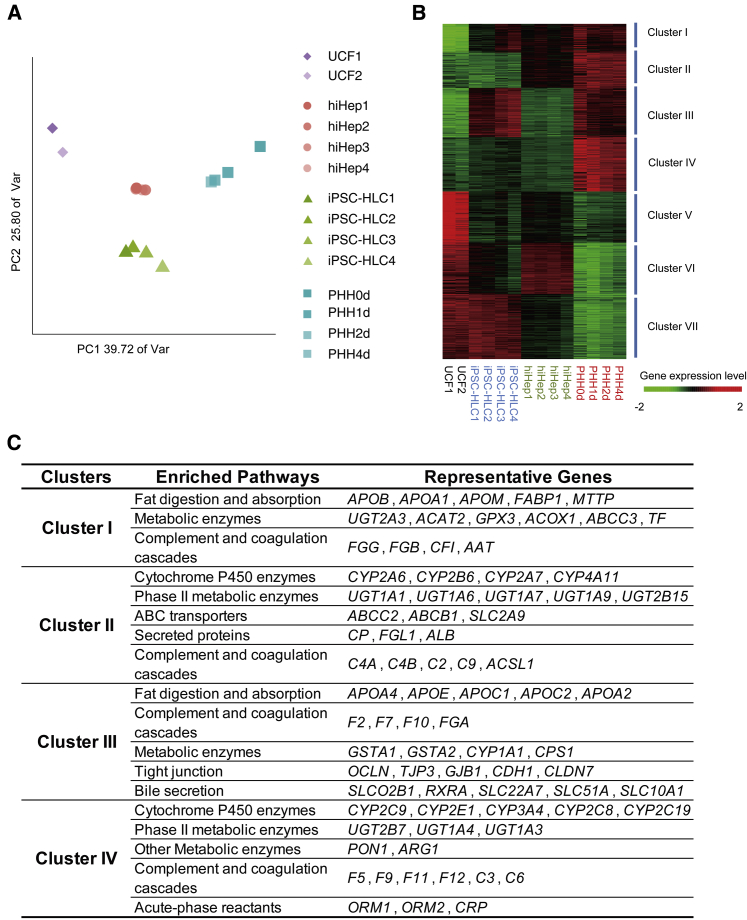
Figure 2.
Transcriptome Analysis of hiHeps and iPSC-HLCs
(A) Principal component analysis (PCA) of four cell types using 4,000 genes with highest variance in UCFs and PHHs cultured for 1, 2, and 4 days. The percentages on the axes represent the variance explained by the respective axes. hiHep1 and hiHep2 were derived from UCF1, hiHep3 and hiHep4 were derived from UCF2; iPSC-HLC1 and iPSC-HLC2 were derived from iPSC1, iPSC-HLC3 and iPSC-HLC4 were derived from iPSC2. PHHs were fresh, or cultured for 1, 2, and 4 days.
(B) Hierarchical clustering of UCFs, hiHeps, iPSC-HLCs, and PHHs using 4,000 genes with highest variance in UCFs and PHHs cultured for 1, 2, and 4 days. The samples are the same as (A).
(C) Enriched pathways and representative genes in different cluster groups are summarized.