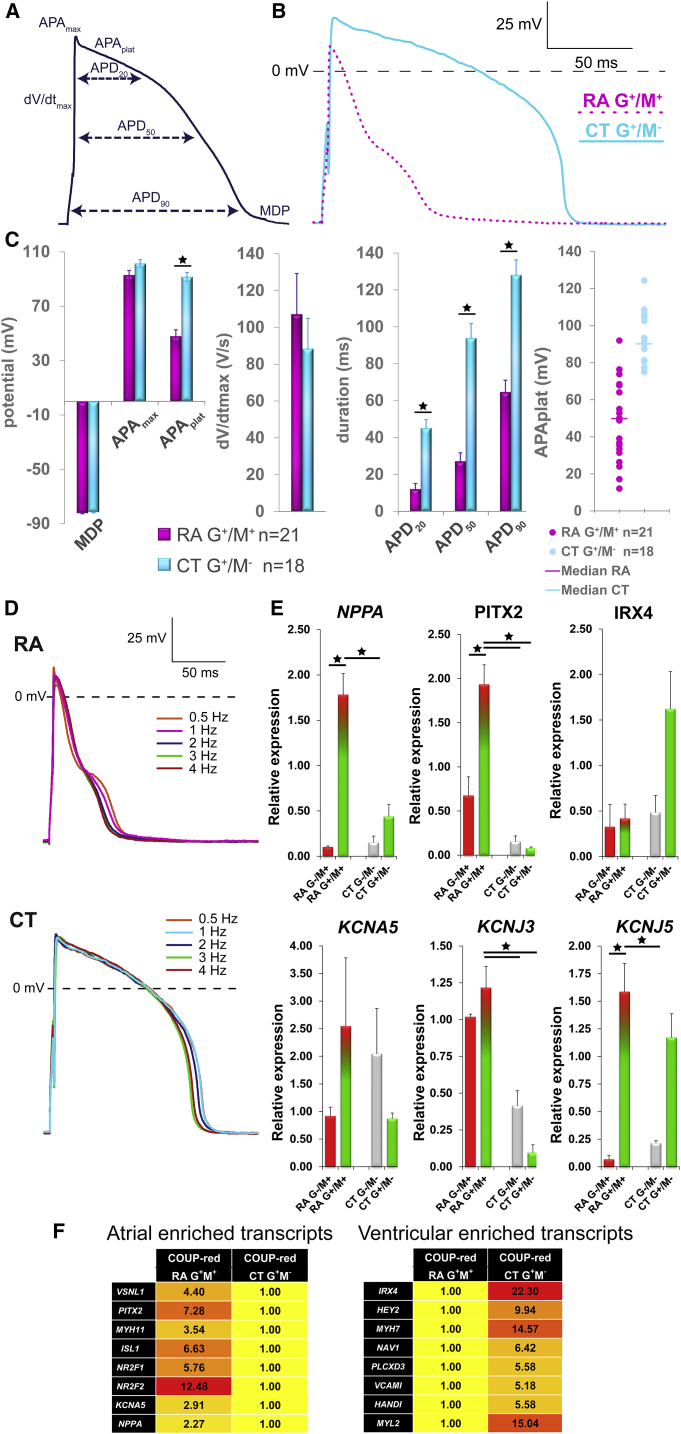
Figure 3.
M+ COUP-red CMs Exhibit Atrial Properties
(A) Analyzed AP parameters.
(B) Representative AP of G+/M+ CMs generated from RA-treated and G+/M− CMs from CT differentiations at 1 Hz stimulation.
(C) Averaged maximum diastolic potential (MDP), maximum AP amplitude (APAmax) and AP plateau amplitude (APAplat), maximum upstroke velocity (dV/dtmax), and AP duration at 20%, 50%, and 90% repolarization (APD20, APD50, and APD90). Scatterplot depicting the APAplat of single CMs and calculated median from RA and CT conditions. n = 21 cells for RA G+/M+ and n = 18 cells for CT G+/M−; from 3 independent differentiations. Data are displayed as means ± SEM; ∗p < 0.05.
(D) Typical AP traces stimulated at different frequencies.
(E) Transcriptional profiling of selected atrial or ventricular-specific genes by qPCR of G+/M+ and G−/M+ fractions from RA differentiations compared with G+/M− and G−/M− fractions from CT differentiations at D20 of differentiations (n = 4). Data are displayed as means ± SEM; ∗p < 0.05.
(F) Genome-wide transcriptional profiling by microarray. Heatmaps to display the fold difference of selected atrial- and ventricular-enriched transcripts in RA-treated G+/M+ and CT G+/M− from COUP-red at a threshold of 2-fold difference.
See also Figure S3.