Figure 6. Consensus EF-Tu demonstrates highly dynamic behavior.
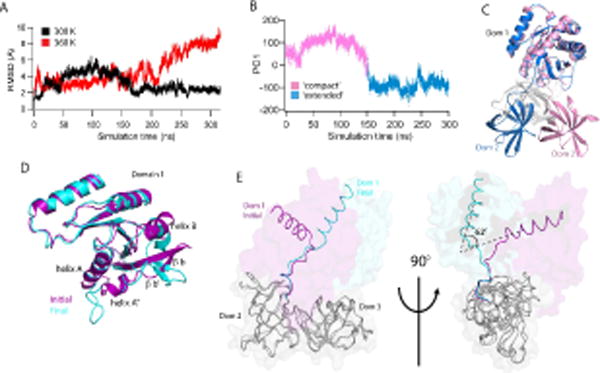
(A) Global RMSDs of the consensus EF-Tu over the simulation, 300 K (black) and 360 K (red). (B) Principal component analysis of Consensus 300 K simulation. First principle component mode (PC1) is the unwinding of the Domain1-Domain2 hinge peptide from a ‘compact’ state to an ‘extended’ state where the domains (1 and 2) are rotated away from each other. Two distinct conformations are observed. (C) Representative structures from each major subspace of PC1. Domain 1 of each conformation is superposed to show unwinding in the hinge peptide from the initial, compact conformation (pink) to the open, extended conformation (blue). Domain 3 is colored in gray for clarity. (D) Local structural changes in consensus 360 K simulation lead to increased RMSD of domain 1. Initial (purple) and final (cyan) structures of the simulation are superposed. Changes include rotation of helix B and unraveling of helices A, A″ and beta strands b′, b. (E) Initial and final conformations of consensus EF-Tu from 360 K simulation are overlaid with domains 2 and 3 (grey) superposed to highlight rotation of domains 2+3 relative to domain 1 (~ 62° rotation). Observed 360 K global RMSD changes result largely from this rotation.
