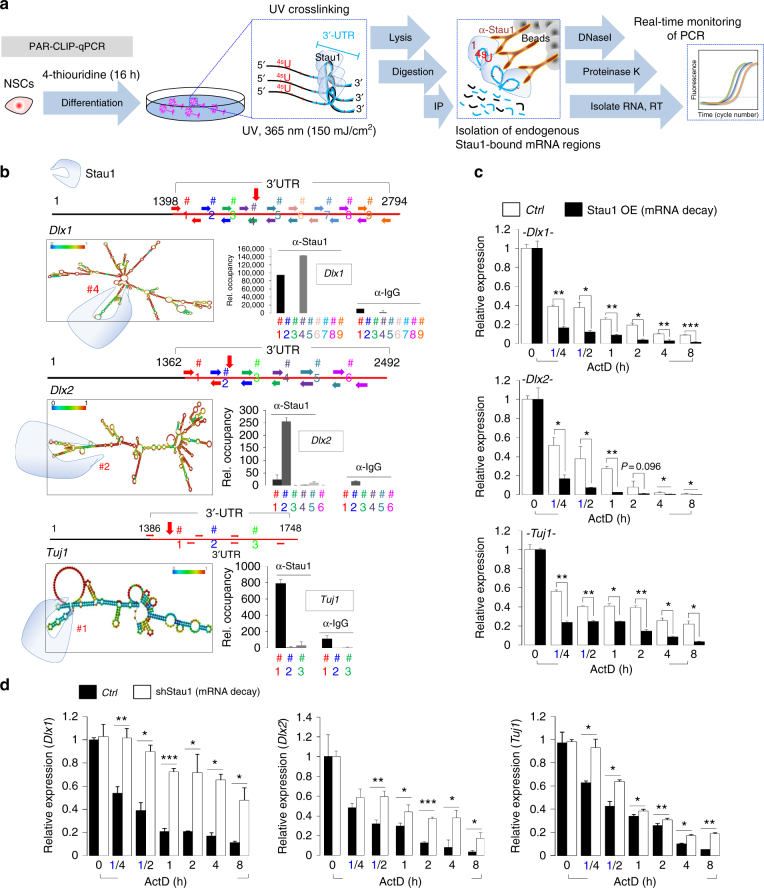
Fig. 5.
Stau1 binds to neurogenesis-associated mRNAs and promotes their degradation. a Schematic showing use of PAR-CLIP-qPCR to detect Stau1 target mRNAs and binding sites. b (Left) RNA secondary structure prediction using online software (http://rna.tbi.univie.ac.at/) of hairpin formation in 3′-UTR sequences of Dlx1, Dlx2, and Tuj1. Potential Stau1 binding regions are marked with vertical red arrows. (Right) PAR-CLIP qPCR shows enrichment of Stau1 binding at various locations on indicated mRNAs. X-axis, primer sets; y-axis, relative fold change. c, d NPCs stably overexpressing (OE) Stau1 or control vectors (c) or infected with shScramble or shStau1 lentiviral vectors (d) were treated with actinomycin D (5 μg/ml) for indicated times and mRNAs were prepared for RNA stability assays (n = 3). Results were normalized to control NPCs at 0 h. Data are presented as mean ± SD. t tests were conducted to calculate statistical significance (*P < 0.05, **P < 0.005, ***P < 0.0005). See also Supplementary Fig. 9a, d and 10a–c