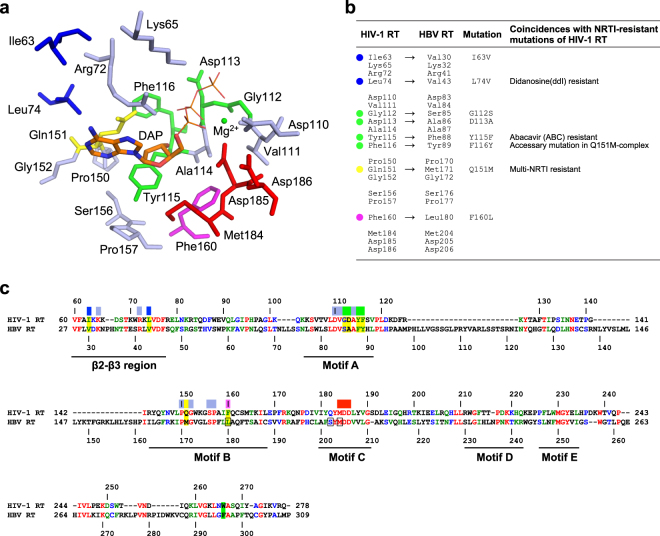
Figure 1.
Design of HIV-1 RT mutants mimicking the N-site of HBV RT in this study. (a) The residues lying within 7 Å of the deoxyribonucleoside moiety of the bound NRTI/dNTP determined by previously reported ternary complexes of HIV-1 RT. This diagram was drawn using the structure of HIV-1 RTWT in complex with DNA and dATP (PDB code, 5TXL)11. The deoxyadenosine moiety of the bound dATP (DAP) is shown in stick model. The unconserved 8 residues selected for mutational analysis are colored in blue (Ile63 and Leu74), green (Gly112, Asp113, Tyr115 and Phe116), yellow (Gln151) and magenta (Phe160). (b) The 20 selected residues in HIV-1 RT and the corresponding residues in HBV RT. The 8 residues selected for mutational analysis are marked with the same color scheme as in (a). Coincidences with NRTI resistance of HIV-1 RT are also indicated. (c) Amino acid sequence alignment between HIV-1 RT and HBV RT. HIV-1 and HBV RT domains were extracted and used for the alignment. The identical, strongly similar, and weakly similar residues are colored in red, green, and blue, respectively. The residues selected for mutational analysis in this work are highlighted in yellow. The conserved motifs (β2-β3 and motifs A, B, C, D, and E) are indicated. The color bars above the sequence indicate the 20 selected residues with the same color scheme as in (a). The residues involving ETV resistance in HBV RT are boxed. Residue stacking at the ribose ring of primer DNA at the N + 3 position shown in Fig. 6 is highlighted in green.