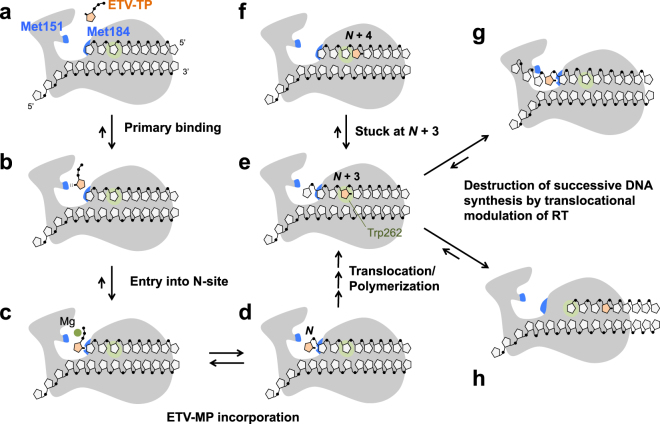
Figure 6.
Proposed model for the mechanism of HIV-1/HBV RT inhibition by ETV-TP. Two Met residues of HIV-1 RT (Met151 and Met184) are labeled and colored in blue (a). ETV-TP is transiently trapped at Met151 (b), then enters into the N-site for tight binding, accepted by relocation of the Met184 side-chain, which is the state ready for ETV-MP incorporation (c). RT catalyzes the ETV-MP addition to the primer DNA 3′-end (d), and successive DNA synthesis continues. When ETV-MP is located at the N + 3 position, strong pausing of DNA synthesis occurs probably owing to DNA strand distortion that leads to 3′-end misalignment at the N-site (e and f). Trp262 of HIV-1 RT is located at the N + 3 position for stacking interactions with the ribose ring. The corresponding residue is substituted with Phe in HBV RT (Phe296). Successive DNA synthesis is finally stopped by translocational modulation of RT, leading to sliding over several nucleotides (g and h). The locations of the slipped DNA strand in (g) and (h) are drawn based on the results of RNase H cleavage analysis reported by Tchesnokov et al.16.