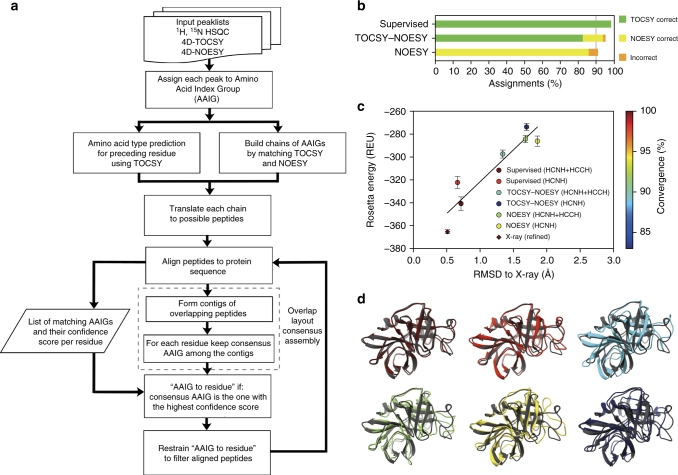
Fig. 3.
Automated structure determination using 4D-CHAINS/autoNOE-Rosetta. a Flowchart of the 4D-CHAINS algorithm for automated NMR resonance assignment from two 4D spectra (TOCSY and NOESY). b Quality of 4D-CHAINS assignments for supervised, TOCSY–NOESY, and NOESY settings, expressed as the average for the four different protein targets. c, d Performance of different 4D-CHAINS assignment scenarios for a 198 aa protein, α-lytic protease, calculated using autoNOE-Rosetta. c Goodness of structural ensembles is measured using the Rosetta all-atom energy function, backbone heavy atom RMSD to X-ray structure (PDB ID 1P01) and degree of structural convergence. Average energy values (in Rosetta Energy Units (REU)) for ensembles calculated using indicated data/assignment scenarios, with errors bars shown at 1 standard deviation. Also shown is the average energy of 10 locally refined X-ray structures (diamond)—refinement adapts the X-ray structure to the local optimum of the Rosetta energy, with a minimum change in RMSD (0.5 Å). The color of points represents the average % of converged residues in each ensemble, according to the color scale on the right. d Lowest-energy structures in each ensemble (shown in the same color as the points in c) superimposed on the X-ray reference structure (gray). Images of structures were produced using Chimera (https://www.cgl.ucsf.edu/chimera)