ABSTRACT
As the notion of small molecule targeting of regulatory viral and cellular RNAs gathers momentum, understanding their structure, and variations thereof, in the appropriate biological context will play a critical role. This is especially true of the ∼1100-nt polyadenylated nuclear (PAN) long non-coding (lnc) RNA of Kaposi's sarcoma herpesvirus (KSHV), whose interaction with viral and cellular proteins is central to lytic infection. Nuclear accumulation of PAN RNA is mediated via a unique triple helical structure at its 3′ terminus (within the expression and nuclear retention element, or ENE) which protects it from deadenylation-dependent decay. Additionally, significant levels of PAN RNA have been reported in both the cytoplasm of KSHV-infected cells and in budding virions, leading us to consider which viral and host proteins might associate with, or dissociate from, this lncRNA during its “journey” through the cell. By combining the power of SHAPE-mutational profiling (SHAPE-MaP) with large scale virus culture facilities of the National Cancer Institute, Frederick MD, Sztuba-Solinska et al. have provide the first detailed description of KSHV PAN nucleoprotein complexes in multiple biological contexts, complementing this by mapping sites of recombinant KSHV proteins on an in vitro-synthesized, polyadenylated counterpart.
KEYWORDS: Kaposi's sarcoma herpes virus; long non-coding RNA; expression and nuclear retention element; in vivo SHAPE-MaP, RNA structure
Background
Kaposi's sarcoma herpesvirus (KSHV), or human herpesvirus 8, is the etiological agent of Kaposi's sarcoma (KS), Primary Effusion Lymphoma (PEL) and Multicentric Castleman's Disease (MCD). In the HAART, era KS remains the second most frequent tumor in HIV-infected patients worldwide, and has become the most common cancer in Sub-Saharan Africa. Strikingly, in the HIV-uninfected population, KSHV seroprevalence ranges from 11% – 69% in Africa, from 4% – 20% in Europe and the Mediterranean, and from 5% – 10% in Asia.7 Despite advances in the treatment of KS, MCD, and PEL, improved therapeutic modalities are needed, and in particular for KS in resource-limited regions.
KSHV produces a highly abundant 1100-nt polyadenylated nuclear (PAN) lncRNA that interacts with both viral and cellular factors as a means of dysregulating expression of genes that modulate the immune response. Given the increased evidence for multifunctionality of lncRNAs in malignant diseases, mechanistic aspects of this understudied species are receiving increased attention. Deciphering their structure, in combination with inter- or intramolecular interactions that mediate their regulatory roles, can be considered the first step in a program of developing RNA- targeted therapeutics. KSHV PAN RNA presents an interesting challenge inasmuch as while the majority is located in the nucleus, significant levels of this lncRNA have been detected in the cytoplasm and, to a lesser extent, in the budding virion2,23 (Fig. 1). Whether extra-nuclear PAN RNA simply reflects “leakage” or discrete nucleoprotein complexes with a specific function dictated by their biological compartment remains to be established. As a first step in this direction, Sztuba-Solinska et al. have performed a detailed structural analysis of nuclear, cytoplasmic and virion-associated KSHV PAN RNA in both the absence and presence of cellular factors, complementing this with an in vitro analysis of PAN RNA/protein complexes.29
Figure 1.
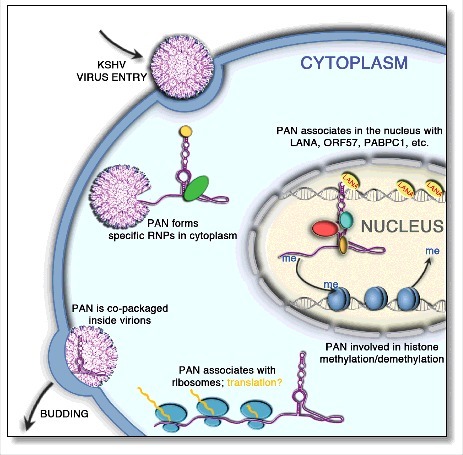
Schematic representation of PAN lncRNA distribution and potential roles in the KSHV lifecycle. Although PAN RNA is considered as primarily a nuclear-associated transcript, previous studies indicated its presence in both the cytoplasm and in virions.2,23 PAN RNA has been shown to associate with (a), translating ribosomes1 (b), histone methylases and demethylases that specifically add and remove the methyl groups22,23 and (c) with various host and viral factors, including ORF57 protein (otherwise referred to as mRNA transcript accumulation factor, or Mta), the polyadenylate-binding protein 1, (PABPC1), and the latency-associated nuclear antigen (LANA).3,16
Verifying biological compartmentalization of PAN
The “gradient” of KSHV PAN lncRNA copy number, varying from ∼500,000 in the nucleus, ∼100,000 in the cytoplasm and several thousand in the budding virion (Fig. 2), presented two significant technical challenges, namely (1) preparing and purifying sufficient amounts of virus for chemical probing and (2) verifying the integrity of nuclear and cytoplasmic RNA preparations. The former benefitted from large scale culture facilities of the AIDS and Cancer Virus Program, National Cancer Institute, Frederick MD, where virions were isolated from a 20-liter, sodium butyrate-induced culture of KSHV-infected BCBL-1 cells by continuous flow, sucrose density gradient ultracentrifugation. The 4-fold difference in MALAT1 lncRNA expression between cytoplasmic and nuclear fractions assessed via quantitative RT-PCR, as well as the absence of peak corresponding to tRNA species in nuclear fraction confirmed the effectiveness of the fractionation.
Figure 2.
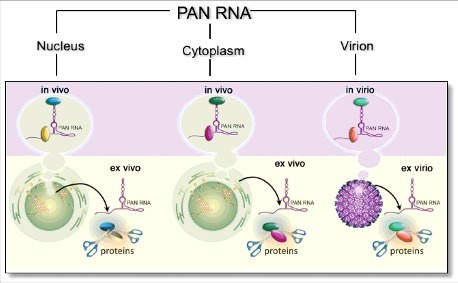
Representation of three biological contexts under which KSHV PAN structure was determined, namely nuclear, cytoplasmic and virion-associated, in both the absence (ex vivo, ex virio) and presence (in vivo, in virio) of cellular factors, which can include both nucleic acid and protein.
RNA structure determination by SHAPE-MaP
Applying the chemoenzymatic probing strategy, selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP), a comprehensive insight has emerged on the modular makeup of PAN RNA. In this technique, RNA is exposed to an acylating reagent that selectively modifies unpaired nucleotides,26,27 after which it is reverse transcribed under conditions in which modified nucleotides are likely to produce a mutation at the complementary position in the cDNA product. cDNA libraries are amplified by PCR and subjected to deep sequencing on an Illumina system. The collective mutation frequencies at individual nucleotide positions of the probed RNA, or mutational profile, directly reflect nucleotide reactivity toward the acylating reagent and therefore the likelihood that it is single or double stranded. This information is used in RNA secondary structure prediction algorithms to assemble maximum likelihood models of RNA structure.
SHAPE-MaP offers numerous advantages compared to other RNA probing techniques26,27). The acylating agent, 1-methyl-7-nitroisatoic anhydride (1M7) is cell permeable, allowing the secondary structures of intracellular (or intra- virion) RNAs to be interrogated. In addition, since the cDNA library can be amplified using gene- specific primers, the sensitivity of SHAPE-MaP far exceeds that of other methods. Quantitation of SHAPE-MaP results is very precise, given the binary nature of mutagenesis and the fact that mutations can easily be counted and compared to the total number of sequence reads. This precision also allows calculation of variability in reactivity values and, consequently, Shannon entropies, which inversely correlate with the frequency that an RNA motif is present among conformers in a population.15,21 Regions of RNA sequence with high Shannon entropies are likely to sample many different conformations, while those with low Shannon entropies are expected to exist in a single conformational state. In general, nucleotides within RNA motifs whose structure is related to its function are found to have low reactivity values and Shannon entropies.17,25
Structural modularity of PAN RNA in various biological contexts
SHAPE-MaP has allowed Sztuba-Solinska et al., to provide the first comprehensive insight into structural modularity of a viral lncRNA during its “journey” through cellular and viral environments. The studies revealed that PAN RNA maintains a branched secondary structure, comprising three domains (Domains I-III, Fig. 3) independent of the biological context. The distribution of individual motifs (helices, stems, internal loops and junctions) correlated well with that estimated for previously described lncRNAs such as HOTAIR and SRA.20,28 Domains I and III were the most compact due to extensive duplex formation, characterized by low SHAPE reactivity and Shannon entropy, strengthening their functional relevance. Domain II was characterized by a flexible conformation, which was proposed to (1), accommodate long-range tertiary interactions (e.g. formation of the ENE triple helix), (2), support more compact folding in adjacent regions and (3) provide an accessible “landing pad” for protein interactions. These data are in stark contrast to transcriptome-wide analysis of messenger RNA showing that they are rather unstructured inside the cell, likely due to processing by RNA helicases, single-stranded RNA binding proteins and other factors that unwind RNAs.24 LncRNAs convey their function mainly through structure, thus it is not surprising that PAN RNA retains its conformation in all biological contexts. Notably, despite PAN sequence heterogeneity noted among KSHV-positive cell lines (VG-1 and BCBL-1) and Kaposi's sarcoma skin lesions of 15 Zambian patients, the structured regions of PAN RNA were conserved among patient isolates, emphasizing their biological importance.
Figure 3.
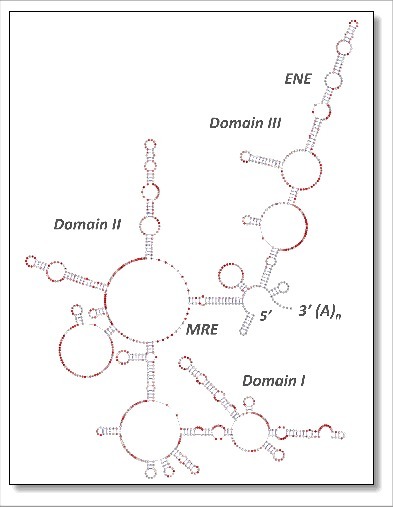
Architecture of the 1100-nt KSHV PAN lncRNA. The secondary structure model is color- coded according to SHAPE-MaP reactivity, where red notations predict single-stranded regions. PAN structure has been arbitrarily depicted as three independent domains (designated Domain I, II and III), within which the cis-acting motifs MRE and ENE are indicated. A significant impediment to reverse transcription proximal to the ENE was interpreted as a stable tertiary interaction such as would be predicted by triple helix formation.5,19 Modified from.29
Conformational differences among nuclear, cytoplasmic and viral PAN RNA were mostly noted for Domain II, and included helical rearrangements around a six-way junction. None of the changes affected the previously characterized cis-acting motifs shown to be essential for PAN RNA stability and function, i.e., the ENE and MRE. Differences in the native structure of PAN RNA may originate from posttranscriptional modifications, of which N6 methyladenosine (m6A) is the most common and previously shown to affect the structure and function of mRNA and lnc RNAs.12,30 At least two potential consensus target sequences for m6A modification on PAN RNA were identified, which may be tip of the “epitranscriptome iceberg” that modulates lncRNA structure and function.9
PAN RNA as a scaffold for distinct protein interactions
The interesting finding with respect to PAN RNA functionality was revealed by difference SHAPE-MaP analysis (ΔSHAPE-MaP), which contrasted reactivity values obtained from ex vivo and ex virio experiments (proteins gently removed, native fold retained) with those obtained in cellullo and in virio, respectively. Using this strategy Sztuba-Solinska et al. identified discrete protein-RNA contact sites that varied among nuclear, cytoplasmic and viral fractions. These findings provided additional layer to PAN RNA structural and functional complexity, as we now realize this essential viral lncRNA not only retains its complex conformation, but also provides a scaffold for a multitude of ribonucleoprotein (RNP) contacts.
Further analysis using in vitro transcribed polyadenylated PAN RNA and individually expressed KSHV ORFs (a unique collection of proteins that have been cloned, expressed and purified at the Frederick National Laboratory for Cancer Research) allowed assignment the identified sites to a selection of protein partners.10 One particularly intriguing result indicated that ORF57, also referred as the mRNA transcript accumulation protein (MTA), binds at the MRE core and is exclusive to nuclear PAN. ORF57 is a potent posttranscriptional regulator that exhibits predominantly nuclear localization and promotes the expression of viral genes, including the three major functions of enhancement of RNA stability, promotion of RNA splicing, and stimulation of protein translation.13,14 Thus, our finding was consistent with previous studies demonstrating that ORF57 is itself primarily nuclear, supporting the notion that ORF57-MRE binding contributes not only to PAN RNA stability,16 but also to nuclear retention. Consistent with a previously proposed role for PAN RNA-Latency Associated Nuclear Antigen (LANA) association, which promotes dissociation of LANA from viral episome,3 multiple LANA contacts sites were revealed, including a small hairpin in Domain II, that overlapped with the protein contact identified only for nuclear PAN RNA. LANA spirally coats the KSHV episome and host DNA via sequence specific and non-specific interactions, forming speckles that contain a plethora of cellular molecules.8 It has been suggested that the association of PAN RNA with LANA promotes dissociation of LANA from viral episome and sequesters the protein during lytic reactivation3 The DNA polymerase processivity factor, ORF59, a cytoplasmic-nuclear shuttling protein18 was also shown to bind PAN RNA, and its contact sites overlapped with those detected in cytoplasmic and nuclear RNA fractions. Finally, minor capsid protein, ORF26 was demonstrated to interact with the ENE, raising the intriguing notion that this interaction could mediate selective packaging of PAN RNA into virions. Taking into account the fact that viral RNA packaging by KSHV11 and murine gammaherpesvirus-69 (MHV-68)4 do not correspond to mRNA expression levels in infected cells, the specific interaction between a structural protein and a cis-acting motif would distinguish PAN RNA from other cellular RNAs during the assembly process. ΔSHAPE- MaP analysis has thus supplied a comprehensive structural and functional roadmap of PAN RNA that will serve further to decipher the involvement of this essential viral lnc RNA in molecular processes that govern KSHV pathogenesis.
Outlook
Structural analysis of RNA has without doubt benefitted immensely from improved and simplified methods of chemoenzymatic probing. However, most studies have been restricted to RNAs derived by in vitro transcription, revealing minimal information on the dynamics of RNA and cellular factors with which they interact. For KSHV PAN, a central player in lytic infection, its presence in multiple biological compartments provides a formidable challenge to structural analysis and at the same time the opportunity to assess topological alterations this lncRNA undergoes as it transits from the nucleus to the budding virion. Several important questions arise immediately from our study. For example, is there a structurally “privileged” PAN RNA subset that is packaged into the virion? Given the importance of posttranscriptional modification, does this play a role in PAN RNA localization? Lastly, the structural modularity noted for PAN RNA in different biological environments is based on 2-dimensional “connectivity” maps and does not shed light on long-range interactions that might be intimately related to PAN function. We see the present work as providing a baseline for such future work.
In addition to highlighting protein binding sites in these biological compartments, we have also taken advantage of the collection of purified KSHV ORFs (curated by AIDS and Cancer Virus Program of the National Cancer Institute) to define binding sites of a subset of these to an in vitro transcribed version of PAN.10 This could be particularly relevant for the capsid protein, ORF26, encoded by a late lytic gene. The demonstration that ORF26 localizes to the ENE may provide a therapeutic opportunity by destabilizing this interaction. Although the mechanistic basis remains to be established, small molecule microarray screening against PAN RNA triple helix has revealed a small molecule that induces lytic reactivation of KSHV following binding to the ENE, raising the possibility that the interaction with ORF26 is compromised (Sztuba-Solinska et al., manuscript in preparation). Targeting the PAN ENE triple helix will allow combinations of lytic activating agents and late lytic cycle inhibitors to be explored as a KSHV “kick-and-kill” strategy.6
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed
Acknowledgements
S.L.G. and J.S.-S. are supported by the Intramural Research Program of The National Cancer Institute, National Institutes of Health, Depertment of Health and Human Services.
References
- 1.Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, Holdorf M, Weissman JS, Ganem D. KSHV 2.0: a comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014;10:e1003847. doi: 10.1371/journal.ppat.1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bechtel J, Grundhoff A, Ganem D. RNAs in the virion of Kaposi's sarcoma-associated herpesvirus. J Virol. 2005;79:10138-46. doi: 10.1128/JVI.79.16.10138-10146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell M, Kim KY, Chang PC, Huerta S, Shevchenko B, Wang DH, Izumiya C, Kung HJ, Izumiya Y. A lytic viral long noncoding RNA modulates the function of a latent protein. J Virol. 2014;88, 1843-48. doi: 10.1128/JVI.03251-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cliffe AR, Nash AA, Dutia BM. Selective uptake of small RNA molecules in the virion of murine gammaherpesvirus 68. J Virol. 2009;83:2321-26. doi: 10.1128/JVI.02303-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conrad NK, Steitz JA. A Kaposi's sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 2005;24:1831-41. doi: 10.1038/sj.emboj.7600662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dissinger NJ, Damania B. Recent advances in understanding Kaposi's sarcoma- associated herpesvirus. F1000Res. 2016. doi: 10.12688/f1000research.7612.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goncalves PH, Uldrick TS, Yarchoan R. HIV-associated Kaposi sarcoma and related diseases. AIDS. 2017;31(14):1903–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hellert J, Weidner-Glunde M, Krausze J, Lunsdorf H, Ritter C, Schulz TF, Luhrs T. The 3D structure of Kaposi sarcoma herpesvirus LANA C-terminal domain bound to DNA. Proc Natl Acad Sci U S A. 2015;112:6694-99. doi: 10.1073/pnas.1421804112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy EM, Courtney DG, Tsai K, Cullen BR. Viral Epitranscriptomics. J Virol. 2017;91(9). doi: 10.1128/JVI.02263-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labo N, Miley W, Marshall V, Gillette W, Esposito D, Bess M, Turano A, Uldrick T, Polizzotto MN, Wyvill KM, et al.. Heterogeneity and breadth of host antibody response to KSHV infection demonstrated by systematic analysis of the KSHV proteome. PLoS Pathog. 2014;10:e1004046. doi: 10.1371/journal.ppat.1004046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin X, Li X, Liang D, Lan K. MicroRNAs and unusual small RNAs discovered in Kaposi's sarcoma-associated herpesvirus virions. J Virol. 2012;86:12717-30. doi: 10.1128/JVI.01473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu N, Pan T. N6-methyladenosine-encoded epitranscriptomics. Nat Struct Mol Biol. 2016;23:98-102. doi: 10.1038/nsmb.3162. [DOI] [PubMed] [Google Scholar]
- 13.Majerciak V, Yamanegi K, Nie SH, Zheng ZM. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J Biol Chem. 2006;281:28365-78. doi: 10.1074/jbc.M603095200. [DOI] [PubMed] [Google Scholar]
- 14.Majerciak V, Zheng ZM. KSHV ORF57, a protein of many faces. Viruses. 2015;7:604-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin JS. Describing the structural diversity within an RNA's ensemble. Entropy-Switz. 2014;16:1331-48. doi: 10.3390/e16031331. [DOI] [Google Scholar]
- 16.Massimelli MJ, Kang JG, Majerciak V, Le SY, Liewehr DJ, Steinberg SM, Zheng ZM. Stability of a long noncoding viral RNA depends on a 9-nt core element at the RNA 5′ end to interact with viral ORF57 and cellular PABPC1. Int J Biol Sci. 2011;7:1145-60. doi: 10.7150/ijbs.7.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mauger DM, Golden M, Yamane D, Williford S, Lemon SM, Martin DP, Weeks KM. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc Natl Acad Sci U S A. 2015;112:3692-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDowell ME, Purushothaman P, Rossetto CC, Pari GS, Verma SC. Phosphorylation of Kaposi's sarcoma-associated herpesvirus processivity factor ORF59 by a viral kinase modulates its ability to associate with RTA and oriLyt. J Virol. 2013;87:8038-52. doi: 10.1128/JVI.03460-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244-47. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novikova IV, Hennelly SP, Sanbonmatsu KY. Sizing up long non-coding RNAs: do lncRNAs have secondary and tertiary structure? Bioarchitecture. 2012;2;189-99. doi: 10.4161/bioa.22592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossetto CC, Pari G. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog. 2012;8:e1002680. doi: 10.1371/journal.ppat.1002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossetto CC, Tarrant-Elorza M, Verma S, Purushothaman P, Pari GS. Regulation of viral and cellular gene expression by Kaposi's sarcoma-associated herpesvirus polyadenylated nuclear RNA. J Virol. 2013;87;5540-53. doi: 10.1128/JVI.03111-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505;701-05. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat Methods. 2014;11:959-65. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smola MJ, Calabrese JM, Weeks KM. Detection of RNA-protein interactions in living cells with SHAPE. Biochemistry. 2015;54:6867-75. doi: 10.1021/acs.biochem.5b00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, Lee DM, Calabrese JM, Weeks KM. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc Natl Acad Sci U S A. 2016;113:10322-27. doi: 10.1073/pnas.1600008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Somarowthu S, Legiewicz M, Chillon I, Marcia M, Liu F, Pyle AM. HOTAIR forms an intricate and modular secondary structure. Mol Cell. 2015;58:353-61. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sztuba-Solinska J, Rausch JW, Smith R, Miller JT, Whitby D, Le Grice SF. Kaposi's sarcoma-associated herpesvirus polyadenylated nuclear RNA: a structural scaffold for nuclear, cytoplasmic and viral proteins. Nucleic Acids Res. 2017;45(11):6805–21. doi: 10.1093/nar/gkx241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou C, Zhang T, Liu F, Zhou J, Ni X, Huo R, Shi Z. The differential expression of mRNAs and long noncoding RNAs between ectopic and eutopic endometria provides new insights into adenomyosis. Mol Biosyst. 2016;12:362-70. doi: 10.1039/C5MB00733J. [DOI] [PubMed] [Google Scholar]


