Figure 3.
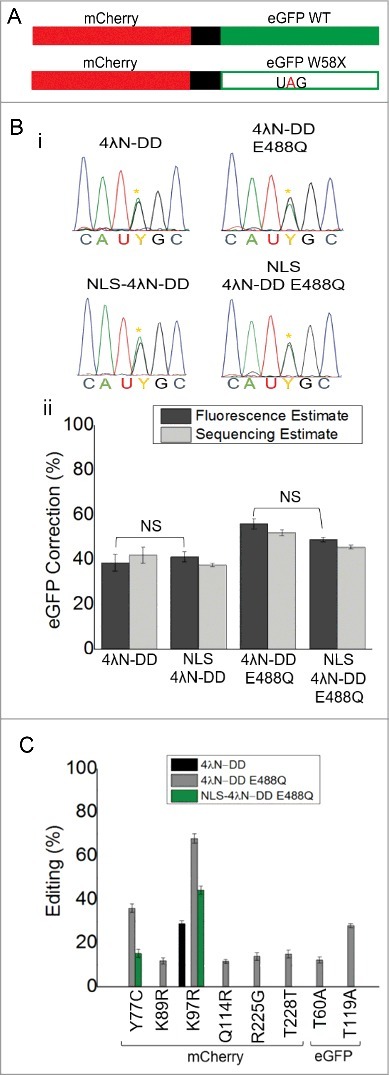
Nuclear localization reduces off-target editing while maintaining on-target editing. (A) Cartoon of fluorescent reporters used for calibration and experimental conditions. Full-length mCherry and eGFP WT were fused together and separated by a 2A self-cleaving peptide shown in black. Another construct was made that had a UAG PTC in eGFP that disrupted green but not red fluorescence (lower panel). (B) Electropherograms from directly sequenced RT-PCR products of experimental cells transfected with all the components of SDRE. Yellow asterisks represent the edited A. (C) Quantification of correction in transfections by both fluorescence (dark grey) and direct sequencing (light grey); Fluorescence estimates were based on n = 200-400 cells and sequencing estimates were taken from the same samples; n = 3, mean ± s.e.m. NS = not significantly different by ANOVA and Tukey's test where p>0.05. (D) Off-target editing events in the complete mCherry-eGFP mRNA transcript. All percentages are based on direct sequencing of RT-PCR products. No off-targets were seen in cells transfected with NLS-4λN-DD and it is not included in the legend; n = 3, mean ± s.e.m.
