Figure 4.
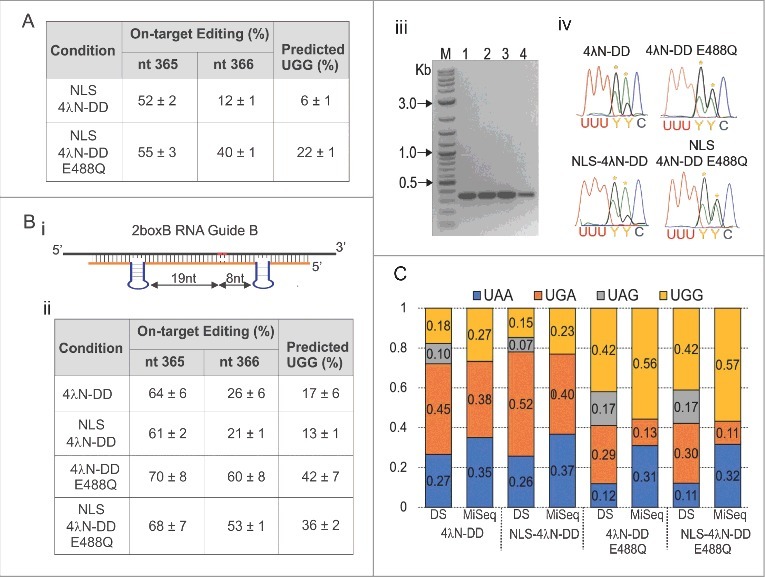
SDRE efficiently corrects a UAA PTC in CFTR Y122X at the level of RNA. (A) Quantification of editing percent of the two A's in the UAA PTC in CFTR Y122X after transfection in HEK293T cells using the NLS-tagged enzymes. Editing estimates were based on RT-PCR products from experimental cells. Percent conversion to UGG for each condition is also shown; n = 3, mean ± s.e.m. (B) An optimized RNA Guide was designed for increasing editing efficiency at the UAA PTC in CFTR Y122X. The optimized RNA guide (2boxB RNA Guide B) was designed with one boxB inserted 19 nt 3′ and another 8 nt 5′ from the second A (i). HEK293T cells were transfected as before followed by RT-PCR and direct sequencing. Quantification of editing estimates at each A and the extent of UGG conversion is shown in (ii). Estimates were based on directly sequenced RT-PCR products from experimental cells. n = 4, mean ± s.e.m. (iii) Image of the 380 bp CFTR amplicon for each condition tested (M = 2-log DNA ladder, 1 = 4λN-DD WT, 2 = NLS-4λN-DD, 3 = 4λN-DD E488Q, and 4 = NLS-4λN-DD E488Q). Bands were extracted and sent for direct sequencing. Examples of electropherograms from the RT-PCR products are shown in (iv). Yellow asterisks indicate the target A's. (C) Conversion to UAG, UGA, UGG, and UAA based on both direct sequencing (DS) and Miseq from the same samples. For the Miseq data, a total of 111,753 transcripts were analyzed in cells transfected with 4λN-DD. For NLS-4λN-DD = 104,011, 4λN-DD E488Q = 92,479 and NLS-4λN-DD E488Q = 113,997.
