Figure 3.
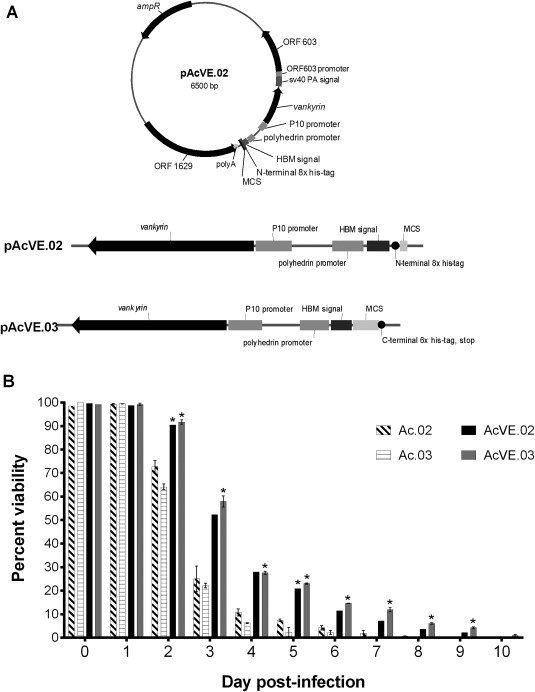
Vankyrin‐encoding baculoviruses increase insect cell viability compared to non‐vankyrin baculoviruses.
Legend: (A) New vankyrin‐encoding transfer vectors pAcVE.02 and pAcVE.03 are shown. Each vector contains an ampicillin resistance gene (ampR), the p10 promoter upstream of the vankyrin gene, a sv40 polyadenylation signal (sv40 PA signal), multi‐cloning site (MCS; pAcVE.02: NotI, SbfI, NheI; pAcVE.03: NcoI, SbfI, XhoI, BstZ17I, BglII, SacII, EagI), honey bee melittin signal peptide (HBM signal), his‐tag, and ORF1629 (including polyA signal) and ORF603 (including promoter)—the two open reading frames that flank the polyhedrin gene in the AcMNPV genome. (B) Viability of SfSWT4 cells infected with baculoviruses generated from transfer vectors pAc.02 and pAc.03, or the vankyrin‐encoding counterpart pAcVE.02 and pAcVE.03. Suspension cultures grown in Sf‐900™II were infected on day 0 with a MOI 5 of each baculovirus. The cell densities of each culture at the time of infections were ∼2 × 106 cells mL−1. Cell viability was determined at the on‐set of the experiment (0 dpi before viral infections) and 1–10 days post‐infection (dpi) by staining cells with trypan blue and counting living cells with the hemocytometer. The data presented are means and standard deviations for duplicate determinations for each infection in a single experiment. The data presented here are representative of multiple experiments performed from which equivalent results were obtained. Statistical significance (P ≤ 0.05) as determined by the Student two‐tailed t test for comparison of non‐vankyrin baculovirus infections vs. vankyrin‐encoding baculovirus infections using the same transfer vector backbone is represented by an asterisk (*).
