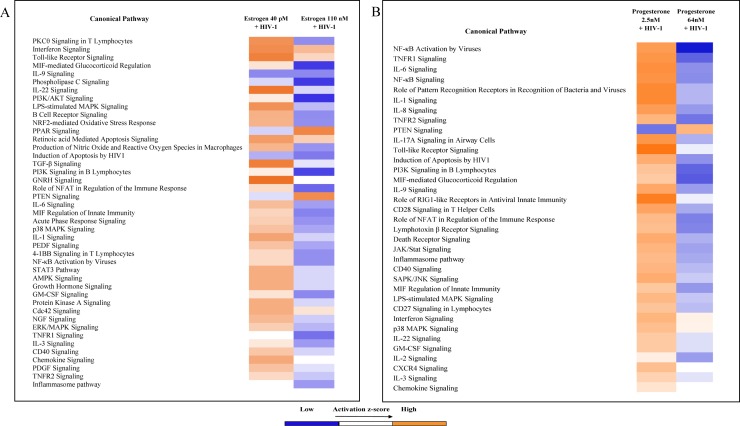
Fig 5. Ingenuity Pathway Analysis (IPA) of the differentially expressed host genes identified by the PCR array.
Data sets were analyzed through the use of QIAGEN's Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, CA, USA www.qiagen.com/ingenuity). Identification of the canonical pathways from Ingenuity Pathways Knowledge Base (IPKB) most significantly associated with the genes differentially expressed between MDMs infected with HIV-1 pre-treated with estrogen (A) or progesterone (B) and MDMs infected with HIV-1 not treated with estrogen or progesterone samples. The differentially expressed genes identified by the PCR array were used for the analysis. The significance of the association was measured on the basis of the ratio of the number of genes from the data set that map to the pathway divided by the total number of genes that map to the canonical pathway (as displayed); and a p-value determining the probability that the association between the genes in the data set and the canonical pathway is explained by chance alone (Fischer’s exact test).