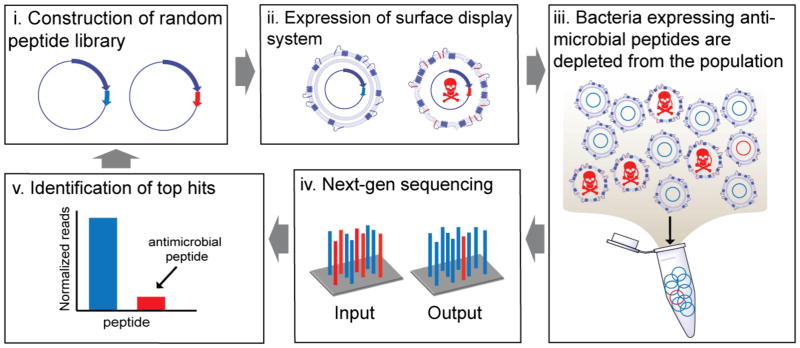
Figure 2. SLAY workflow.
Batch screening of peptides using our surface display system can be achieved by first constructing a random library using random PCR primers that flank the peptide region (i), followed by collection of transformants, plasmid isolation, and subsequent transformation into a bacterial strain of interest. Next, the library is grown in culture and induced (ii). Peptides with antimicrobial activity (colored red) will drop out of the population (iii). Next-generation sequencing of the initial input at time zero and output (iv) at a pre-defined number of hours provides a read out of sequencing counts (v). From this information, top hits can be identified and tested. Further libraries can be constructed based on the identified top hits and the process can be repeated. A more detailed explanation of our workflow can be found in the methods section.