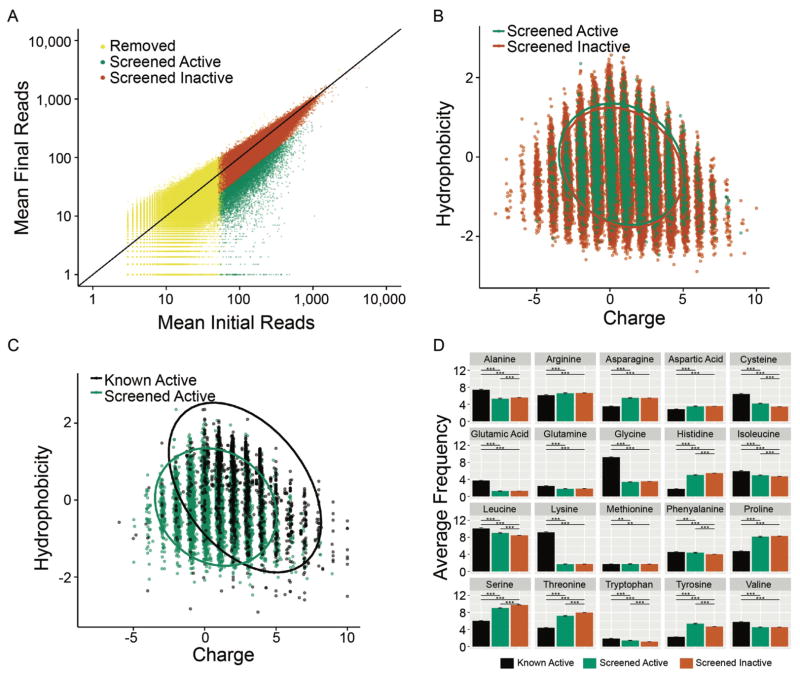
Figure 4. Computational analysis of the random peptide library screen results.
(A) Mean normalized input and output counts of total peptide library. Peptides considered active with lfcMLE < or = −1 are plotted in green. Peptides with lfcMLE > −1 were considered inactive are plotted in orange. Peptides removed from further analysis contained initial reads in either replicate of less than or equal to 50 and are plotted in yellow. (B) Screened peptides are plotted according to their hydrophobicity and charge properties. Active peptides are colored in green and inactive peptides are colored in orange. Ellipses represent a 95% confidence interval assuming a t-distribution. (C) A charge vs hydrophobicity plot comparing SLAY active peptides and known active peptides. Known antimicrobial peptides complied from six available online databases are colored in black, active peptides from our screen are colored green. Ellipses represent a 95% confidence interval assuming a t-distribution. (D) Plot of amino acid frequencies of known, active and inactive peptides from our screen. The error bars represent the SEM (standard error of the mean) and the asterisks correspond to Bonferroni adjusted p-values (*, **, and *** denote p-value <0.05, <0.01, and <0.001 respectively) derived from Tukey’s range test performed in conjunction with an ANOVA.