Figure 3. PGAM5 interacts with AIFM1.
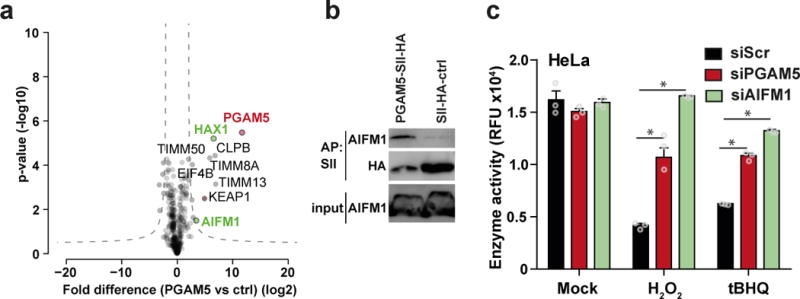
(a) Identification of PGAM5 binding partners. Proteins enriched with SII-HA-PGAM5 and THYN1 (ctrl) from HeLa FlpIn cells were analyzed by LC-MS/MS. Volcano plots show the average degrees of enrichment by PGAM5 over control (ratio of label-free quantitation (LFQ) protein intensities; x-axis) and p-values (two-tailed t-test; y‐axis) for each protein. Significantly enriched proteins (FDR 0.001, S0=0.2) are separated from background proteins by a hyperbolic curve (dotted line). Red: PGAM5, green: proteins known to be involved in cell death. Four independent affinity purifications were performed for both baits. (b) Binding of endogenous AIFM1 to PGAM5. Immunoblot analysis of SII-HA-PGAM5 or SII-HA-ctrl precipitates and input lysate. One representative experiment of eight is shown. (c) Viability of HeLa cells treated with siRNA against PGAM5, AIFM1 and siScr after 21h of H2O2 or tert-Butylhydroquinone (tBHQ) treatment. Cell viability was tested by resazurin conversion assay. The plot shows the mean ± S.D. of three individual treatments. One representative experiment of four is shown. * p-value < 0.001, 2way ANOVA.
