Figure 1.
Design of the peptide-HLA-A*02:01 yeast-display library.
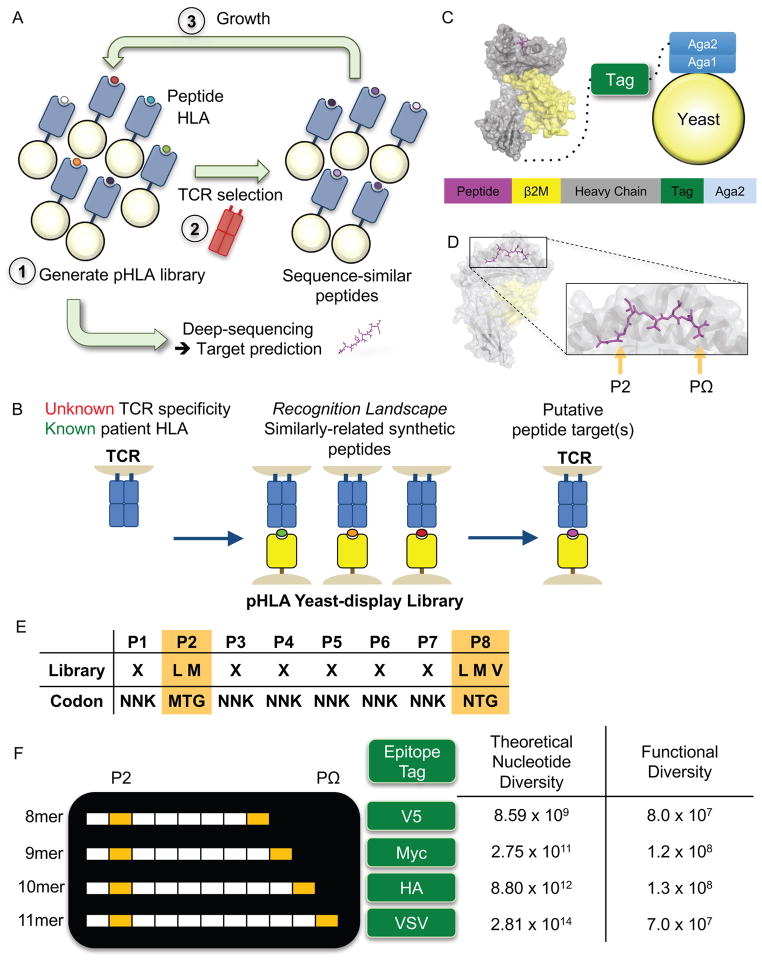
(A) Methodology for selecting a yeast-display library of pHLA. Each yeast displays a unique peptide that is genetically encoded. A typical library contains ~108 unique peptides, which is selected by a TCR of interest. Yeast are enriched in an affinity-based selection using bead-multimerized TCR and grown for iterative rounds of selection. Peptides are successively enriched and all yeast DNA is deep-sequenced. These synthetic peptide sequences are used to generate a model to make predictions for TCR ligands derived from the human proteome and/or patient-specific exome.
(B) The goal of the study is to use the yeast-display selection to de-orphanize a TCR of unknown antigen specificity. The peptides selected by a TCR from the yeast-display selection generates a recognition landscape for a particular TCR, which is then used to make predictions of antigen specificity for orphan TCRs. Predicted targets can be validated in a T cell stimulation assay.
(C) The construct utilizes a single-chain design to display the pHLA-A*02:01 complex tethered to an epitope tag and Aga2p, which binds to the native Aga1 protein on yeast. Each component is connected covalently by a Gly-Ser linker. The epitope tag is introduced to monitor expression of the library.
(D) The MART-1/HLA-A*02 complex structure (PDB 4L3E) highlighting the two peptide anchors with orange arrows. These peptide positions at P2 and PΩ of the peptide allow for peptide binding to HLA-A*02.
(E) An example 8mer peptide library shows the anchor preferences for the HLA-A*02:01 library and the remaining positions that are randomized to any of the twenty amino acids (X = twenty amino acids and stop codon). Nucleotide abbreviations for codon usage are listed according to the IUPAC nucleotide code.
(F) A multi-length library designed to capture the most common length peptides presented by HLA-A*02:01. Each peptide length is placed in a construct using a unique epitope tag for selection monitoring. The libraries have theoretical nucleotide diversities dictated by the peptide length and library composition. The functional diversity represents the true capacity of the physical libraries based on yeast colony counting after limiting dilution of the library.