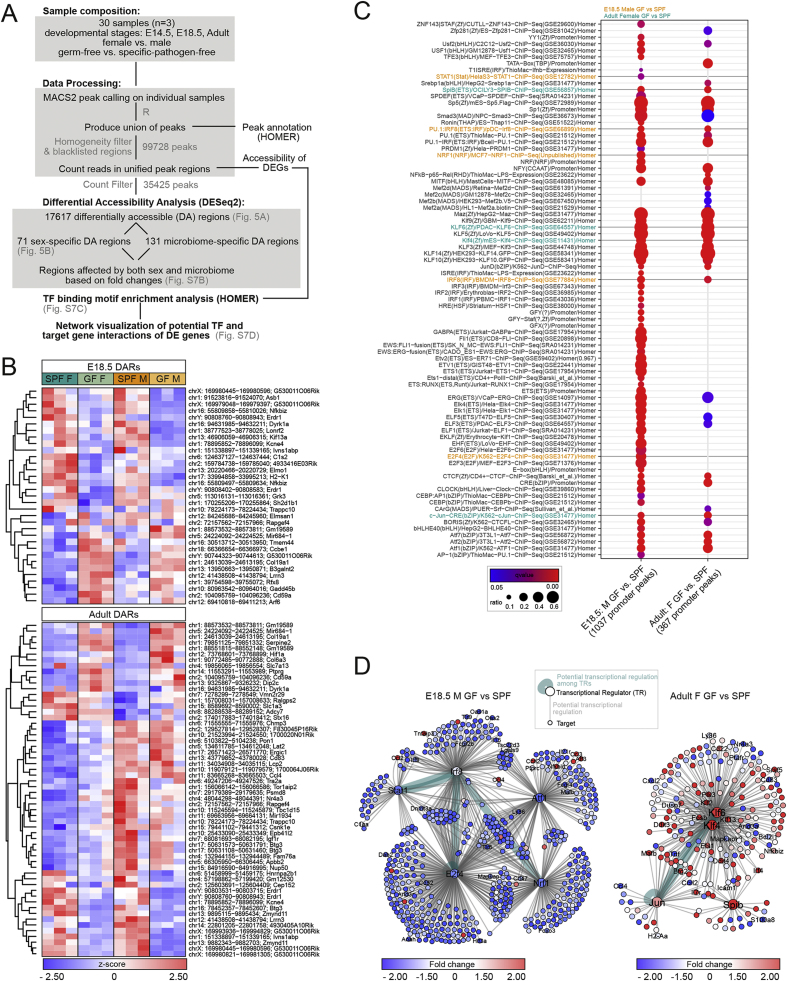
Figure S7.
ATAC-Seq Reveals Temporal Changes in Chromatin Accessibility of Germ-Free Mice, Related to Figure 5
(A) Schema illustrating the workflow of the bioinformatics ATAC-seq analysis in microglia from SPF and GF mice at E14.5, E18.5 and in adult.
(B) Heatmap showing the hierarchical clustering of the DARs (FDR < 0.1) affected by both sex and microbiome with a FC of at least 1.5 due to both factors, colored according to z-transformed read counts (cpm) from blue (low count) to red (high count) in microglia from SPF and GF mice at E18.5 and in adult. n = 3 replicates per condition and stage, with each replicate obtained by pooling microglia from 1 to 3 brains.
(C) Dot plot showing significantly enriched transcription factor binding motifs (q-value < 0.05) in the ATAC-seq peak sequences found in promoter regions of the indicated sets of DEGs. Dot size indicates the ratio of sequences featuring the respective motif to the total number of tested sequences, and dot color illustrates the q-value of the enrichment. Green motifs correspond to transcription factors differentially expressed between GF and SPF male microglia at E18.5 and orange motifs to transcription factors differentially expressed between GF and SPF female adult microglia.
(D) Network visualization of differentially-expressed transcription factors corresponding to enriched binding motifs and their potential target genes among the DEGs between E18.5 male GF and SPF (left panel) and adult female GF and SPF (right panel). Grey edges indicate a potential regulation of the target gene by the transcription factor and turquoise edges present potential regulation between transcription factors. Nodes are colored according to their FC of the indicated comparison.