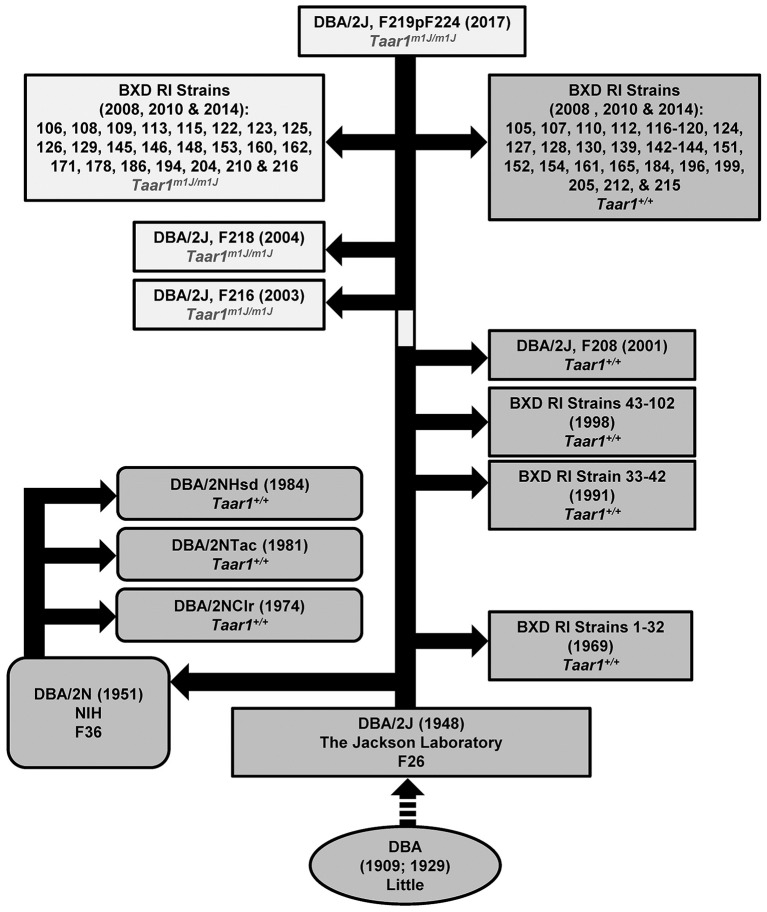
Figure 8.
Chronology for the Taar1m1J allele. The historical sequence for DBA/2 mice is indicated, to the best of our knowledge. Dr. Clarence Cook Little began inbreeding the first DBA strain in 1909. In 1929, The Jackson Laboratory was established by Dr. Little, and strain crosses were initiated from which DBA sublines emerged, including the DBA/2. By 1948, the F26 generation had been produced at The Jackson Laboratory. F36 DBA/2 mice were distributed to the National Institutes of Health (NIH) in 1951, and named DBA/2N. Mice from NIH were distributed to Charles River, Taconic, and Harlan Sprague Dawley (now Envigo) in 1974, 1981, and 1984, respectively, where the mice were renamed to indicate source (DBA/2NClr, DBA/2NTac, and DBA/2NHsd, respectively). Current genotyping of mice sourced from those suppliers indicates that all are homozygous Taar1+. Multiple waves of BXD RI strains have been initiated beginning ca 1969, ca 1991, ca 1998, ca 2008, ca 2010, and ca 2014 (for details see Taylor et al., 1973, 1975, 1999; Peirce et al., 2004; http://www.genenetwork.org/mouseCross.html). DNA samples were available from many strains and Taar1 genotyping results are indicated. Some sequencing results for BXD RI strains not used in the current manuscript have been published (Shi et al., 2016). Only the more recently derived BXD RI strains possess the mutant Taar1m1J allele. Genotyping of DNA from generation F208, F216, and F218 DBA/2J mice indicates that the SNP in Taar1 at position 229 arose in 2001-2003 (indicated by the light coloration on the main bar of the figure). The dashed arrow indicates that some intermediary events have been omitted. Sequencing results for some additional strains can be found in Shi et al. (2016). Note that among strains BXD1-102, the following were directly sequenced for Taar1 position 229 genotype: 1, 2, 5, 6, 8, 9, 11, 12, 14–16, 18–25, 27–34, 36, 38–40, 42–45, 48, 48a, 51, 55, 56, 60, 63–66, 68–71, 73–75, 77, 79, 83, 84, 87, 89, 90, and 98–102.