Abstract
Studies have indicated that thrombophilic genes polymorphisms are associated with recurrent pregnancy loss (RPL) in the Iranian population. We aimed to evaluate the precise association between thrombophilic genes polymorphisms (MTHFR C677T, MTHFR A1298C, Prothrombin G20210A, FVL G1691A, and PAI-1 4G/5G) and RPL risk in the Iranian population. PubMed, Web of Science, Google Scholar, and ISC were searched for eligible articles published up to April 1, 2017. In total, 37 case-control studies in 18 relevant publications were selected: 1,199, 1,194, 630, 830, and 955 RPL cases and 1,079, 1079, 594, 794, and 499 controls for MTHFR C677T, MTHFR A1298C,Prothrombin G20210A, FVL G1691A, and PAI-1 4G/5G, respectively. The results indicated a significant increased risk of RPL in all genetic models in the population. Also, Prothrombin G20210A and FVL G1691A as well as PAI-1 4G/5G polymorphisms were associated with RPL risk in the Iranian population. Hence, thrombophilic genes polymorphisms are associated with an increased RPL risk in the Iranian population.
Keywords: Recurrent miscarriage, Thrombophilia, Factor V leiden, Prothrombin, Meta-analysis
INTRODUCTION
The miscarriage of three or more consecutive pregnancies in the first or early second trimester is termed as recurrent pregnancy loss (RPL)[1]. Several etiological factors, including endocrinologic problems, uterine structural, chromosomal anomalies, and antiphospholipid antibody syndrome can be the causes of some RPL cases. However, in many cases, the pathogenesis of RPL remains unknown[2,3]. For more than two decades, researchers have focused on certain inherited thrombophilic factors that may be the risk of arterial and/or venous thromboses and their possible association with pregnancy complications such as early pregnancy loss[4]. It is estimated that the thrombophilia is a common cause of RPL and is found in 40-50% of cases[5].
Three common inherited thrombophilia markers, namely Factor V Leiden (FVL), Prothrombin G20210A (PT G20210A), and Methylene tetrahydrofolate reductase (MTHFR) C677T are candidate genes for venous thromboembolism (VTE)[5]. Various hypotheses were proposed to explain the role of the thrombophilic genes polymorphisms such as MTHFR C677T, MTHFR A1298C, Prothrombin G20210A, FVL G1691A, and PAI-1 4G/5G in RPL[6,7]. A large number of studies have investigated the association between the thrombophilia gene polymorphisms and RPL susceptibility in the Iranian population[8-25]. However, the results were inconsistent or inconclusive, presumably due to the small sample size in these published studies. Undoubtedly, meta-analysis can be used to increase power and answer questions not posed by the individual studies. Therefore, we conducted this systematic review and meta-analysis to investigate the association between the most common polymorphisms of thrombophilic genes (including MTHFR C677T, MTHFR A1298C, Prothrombin G20210A, FVL G1691A, and PAI-1 4G/5G) and RPL risk in the Iranian population.
MATERIALS AND METHODS
Search strategy
To identify eligible studies for this meta-analysis, we searched the PubMed, Web of Science, Google Scholar databases, ISC, and EMBASE. In the search, we considered all eligible articles published up to April 1, 2017 that examined the association between the MTHFR C677T, MTHFR A1298C, Prothrombin G20210A, FVL G1691A, and PAI-1 4G/5G polymorphisms and RPL risk in the Iranian population. The following key terms were included in our search: “recurrent pregnancy loss”, ‘’recurrent miscarriage’’, ‘’habitual abortion’’, “RPL”, “thrombophilic gene”, “MTHFR C677T’’, “MTHFR A1298C’’, “Prothrombin G20210A’’, “Factor V Leiden G1691A’’, “PAI-1 4G/5G’’, “polymorphism”, “variant”, “gene”, “genotype”, “SNP”, and “allele”. The extracted publications were limited to Persian and English languages and conducted only on human subjects. We retrieved those publications matching the keywords without no restriction, and then the studies were evaluated by reading the title and abstract. We have also screened the reference lists of the retrieved articles for original papers. If there were multiple reports of the same study or overlapping data, only the study with the largest sample sizes or the most recent one was selected in our meta-analysis, and the others were excluded.
Inclusion and exclusion criteria
The included studies to the meta-analysis had to be consistent with the following criteria: (1) published in full-text, (2) be case-control or cohort design, 3) evaluate the association between MTHFR C677T, MTHFR A1298C, Prothrombin G20210A, FVL G1691A, and PAI-1 4G/5G polymorphisms and the risk of RPL in the Iranian populations, (4) offered the size of the sample and sufficient data (genotype distributions of both cases and controls were available) for estimating OR with 95% CI, and (5) written in English or Persian. The exclusion criteria were as follows: (1) abstracts, case reports, letter to the editor, and reviews, (2) studies with only case group (no control population), (3) studies on other poly-morphisms of thrombophilic genes, (4) studies without detail genotype frequencies in which calculation of OR is impossible, and (5) duplicate publications of data from the same study.
Data extraction
Two investigators independently extracted the data using a pre-designed form. Based on the inclusion and exclusion criteria, we extracted the following data from each study: the first author, year of publication, number of RPL patients and controls, genotype and allele frequency, minor allele frequencies (MAFs) in control subjects, and Hardy-Weinberg equilibrium (HWE) test in control subjects. For conflicting evaluation, these two investigators carried out discussions until a consensus was reached.
Statistical analysis
All the statistical analyses were performed by comprehensive meta-analysis (CMA) version 2.0 software (Biostat, USA). All P values were two-tailed with a significant level at 0.05. The strength of associations was assessed by using ORs and 95% CIs, and the significance of pooled ORs was examined by Ztest. We performed a meta-analysis of the association between MTHFR C677T polymorphism and RPL under the allelic model (T vs. C), the homozygote model (TT vs. CC), the heterozygote model (CT vs. CC), the dominant model (TT + CT vs. CC), and the recessive model (TT vs. CT + CC). The MTHFR A1298C polymorphism was evaluated using the allelic model (C vs. A), the heterozygote model (AC vs. AA), the homozygote model (CC vs. AA), the dominant model (CC + AC vs. AA), and the recessive model (CC vs. AC + AA). The Prothrombin G20210A and FVL G1619A polymorphisms were assessed under the allelic model (A vs. G), heterozygote model (GA vs. GG), the homozygote model (AA vs. GG), the dominant model (AA + AG vs. GG), and the recessive model (AA vs. AG + GG). PAI-1 4G/5G polymorphism was assessed under the allelic model (4G vs. 5G), the heterozygote model (4G/5G vs. 5G/5G), the homozygote (4G/4G vs. 5G/5G), the dominant model (4G/4G + 4G/5G vs. 5G/5G), and the recessive model (4G/4G vs. 4G/5G + 5G/5G). Heterogeneity assumption was checked by a chi-square-based Q-test, and I2 statistics was calculated to quantify the proportion of the total variation across studies due to heterogeneity[26]. The heterogeneity was considered significant if either the Q statistics had p < 0.1 or I2 > 50%. An I2 value of 0% represents no heterogeneity, and with the values of 25%, 50%, 75%, or more, it represents low, moderate, high, and extreme heterogeneity, respectively. A P value greater than 0.10 indicated the lack of heterogeneity among studies; therefore, the fixed-effects model (Mantel-Haenszel method) was used to calculate pooled OR[27]. Otherwise, the fixed-effects model (Mantel-Haenszel approach) was used. HWEs were calculated with goodness-of-fit tests (i.e., chi-square or Fisher’s exact tests). A value of p < 0.01 signified a departure from HWE[28]. One-way sensitivity analyses were carried out by consecutively omitting one study at a time to assess the power of the meta-analysis findings[29]. Visual inspection of the asymmetry of funnel plots was carried out to assess potential publication bias. Begg’s funnel plot, a scatter plot of effect against a measure of study size, was generated as a visual aid to detect bias or systematic heterogeneity[30]. Publication bias was assessed by Egger’s test; p < 0.05 was considered statistically significant[31]. Sensitivity analysis was performed to evaluate the stability of the results by removing the studies, but not in HWE.
RESULTS
Characteristics of included studies
Based on the search criteria, 53 individual literatures were found. After screening the titles and abstracts, 35 publications that did not meet the criteria were excluded. These studies were reviews, short reports, case reports, and other polymorphisms of MTHFR, Prothrombin, FVL, and PAI-1 genes. As summarized in Tables 1 and 2, a total of 37 case-control studies in 18 publications[8-25] were selected in the final meta-analysis, including 1,199 RPL cases and 1,079 controls for MTHFR C677T (from ten studies), 1,194 RPL cases and 1079 controls for MTHFR A1298C (from ten studies), 630 RPL cases and 594 controls for Prothrombin G20210A (from five studies), 830 RPL cases and 794 controls for FVL G1691A (from seven studies), and 955 RPL cases and 499 controls for PAI-1 4G/5G (from five studies). Genotype distributions in the controls of ten case-control studies were not in agreement with HWE.
Table 1.
Characteristics of studies included in MTHFR C677T and A1298AC polymorphisms and RPL
| First author | Year | Case/Control | Cases | Controls | MAFs | HWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Allele | Genotype | Allele | |||||||||||
| MTHFR C677T | CC | TC | TT | C | T | CC | TC | TT | C | T | ||||
| Bagheri[8] | 2010 | 61/53 | 34 | 22 | 5 | 90 | 32 | 27 | 21 | 5 | 75 | 31 | 0.292 | 0.756 |
| Jeddi-Tehrani[9] | 2011 | 100/100 | 43 | 42 | 15 | 128 | 72 | 66 | 25 | 9 | 157 | 43 | 0.215 | <0.001 |
| Kazerooni[10] | 2012 | 60/62 | 50 | 6 | 4 | 106 | 14 | 54 | 6 | 2 | 114 | 10 | 0.080 | 0.006 |
| Poursadegh Zonouzi[11] | 2012 | 89/50 | 53 | 30 | 6 | 136 | 42 | 27 | 22 | 1 | 76 | 24 | 0.240 | 0.144 |
| Idali[12] | 2012 | 106/100 | 61 | 36 | 9 | 158 | 54 | 66 | 25 | 9 | 157 | 43 | 0.215 | 0.009 |
| Eskandari[13] | 2013 | 105/98 | 43 | 48 | 14 | 134 | 76 | 61 | 30 | 7 | 152 | 44 | 0.224 | 0.231 |
| Khaleghparast[14] | 2014 | 30/10 | 13 | 17 | 0 | 43 | 17 | 5 | 5 | 0 | 15 | 5 | 0.250 | 0.291 |
| Yousefian[15] | 2014 | 204/116 | 96 | 90 | 18 | 282 | 126 | 63 | 43 | 10 | 169 | 63 | 0.271 | 0.497 |
| Farahmand[16] | 2015 | 330/350 | 180 | 114 | 36 | 474 | 186 | 230 | 85 | 35 | 545 | 155 | 0.221 | <0.001 |
| Najafian[17] | 2016 | 114/140 | 30 | 48 | 36 | 108 | 120 | 58 | 56 | 0 | 172 | 56 | 0.245 | <0.001 |
| MTHFR 1298C | AA | CA | CC | A | C | AA | CA | CC | A | C | ||||
| Bagheri[8] | 2010 | 61/53 | 24 | 28 | 9 | 76 | 46 | 21 | 24 | 8 | 66 | 40 | 0.377 | 0.791 |
| Jeddi-Tehrani[9] | 2011 | 100/100 | 69 | 27 | 4 | 165 | 35 | 94 | 6 | 0 | 194 | 6 | 0.030 | 0.757 |
| Poursadegh Zonouzi[11] | 2012 | 89/50 | 35 | 46 | 8 | 116 | 62 | 13 | 34 | 3 | 60 | 40 | 0.400 | 0.003 |
| Idali[12] | 2012 | 106/100 | 40 | 46 | 20 | 126 | 86 | 94 | 6 | 0 | 194 | 6 | 0.030 | 0.757 |
| Sheikhha[18] | 2012 | 60/60 | 8 | 45 | 7 | 51 | 49 | 34 | 26 | 0 | 94 | 26 | 0.216 | 0.032 |
| Khaleghparast[19] | 2014 | 30/10 | 11 | 13 | 6 | 35 | 25 | 5 | 2 | 3 | 12 | 8 | 0.040 | 0.065 |
| Yousefian[15] | 2014 | 204/116 | 98 | 81 | 25 | 277 | 131 | 68 | 39 | 9 | 175 | 57 | 0.245 | 0.316 |
| Farahmand[16] | 2015 | 330/350 | 134 | 152 | 44 | 420 | 240 | 329 | 20 | 1 | 678 | 22 | 0.031 | 0.250 |
| Arabkhazaeli[19] | 2016 | 100/100 | 100 | 0 | 0 | 200 | 0 | 100 | 0 | 0 | 200 | 0 | 0.00 | 0.250 |
| Najafian[17] | 2016 | 114/140 | 30 | 48 | 36 | 108 | 120 | 58 | 56 | 0 | 172 | 56 | 0.245 | <0.001 |
MAFs, minor allele frequencies; HWE, Hardy-Weinberg equilibrium
Table 2.
Characteristics of studies included in Prothrombin G20210A, FVL G1691A, and PAI-1 4G/5G polymorphisms and RPL
| First author | Year | Case/control | Cases | Controls | MAFs | HWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Allele | Genotype | Allele | |||||||||||
| Prothrombin G20210A | GG | AG | AA | G | A | GG | AG | AA | G | A | ||||
| Bagheri[8] | 2011 | 70/60 | 48 | 22 | 0 | 118 | 22 | 57 | 3 | 0 | 117 | 3 | 0.025 | 0.842 |
| Kazerooni[10] | 2013 | 60/60 | 52 | 8 | 0 | 112 | 8 | 54 | 6 | 0 | 114 | 6 | 0.050 | 0.683 |
| Teremmahi Ardestani[21] | 2013 | 80/80 | 80 | 0 | 0 | 160 | 0 | 80 | 0 | 0 | 160 | 0 | 0.00 | 0.683 |
| Parand[21] | 2013 | 90/44 | 88 | 2 | 0 | 178 | 2 | 43 | 1 | 0 | 87 | 1 | 0.011 | 0.939 |
| Farahmand [16] | 2015 | 330/350 | 316 | 14 | 0 | 646 | 14 | 340 | 10 | 0 | 690 | 10 | 0.014 | 0.786 |
| FVL 1619 G/A | GG | AG | AA | G | A | GG | AG | AA | G | A | ||||
| Bagheri[8] | 2011 | 70/60 | 70 | 0 | 0 | 140 | 0 | 60 | 0 | 0 | 120 | 0 | 0.00 | 0.786 |
| Kazerooni[10] | 2013 | 60/60 | 43 | 12 | 5 | 98 | 22 | 54 | 4 | 2 | 112 | 8 | 0.066 | 0.003 |
| Torabi[22] | 2012 | 100/100 | 87 | 12 | 1 | 186 | 14 | 96 | 4 | 0 | 196 | 4 | 0.002 | 0.833 |
| Teremmahi Ardestani[20] | 2013 | 80/80 | 78 | 2 | 0 | 158 | 2 | 79 | 1 | 0 | 159 | 1 | 0.006 | 0.955 |
| Parand[21] | 2013 | 90/44 | 72 | 15 | 3 | 159 | 21 | 38 | 6 | 0 | 82 | 6 | 0.068 | 0.627 |
| Farahmand[16] | 2015 | 330/350 | 302 | 28 | 0 | 632 | 28 | 340 | 10 | 0 | 690 | 10 | 0.014 | 0.786 |
| Arabkhazaeli[19] | 2016 | 100/100 | 95 | 5 | 0 | 195 | 5 | 91 | 9 | 0 | 191 | 9 | 0.045 | 0.637 |
| PAI-1 4G/5G | 5G5G | 5G4G | 4G4G | 5G | 4G | 5G5G | 5G4G | 4G4G | 5G | 4G | ||||
| Jeddi-Tehrani[9] | 2011 | 100/100 | 60 | 31 | 9 | 151 | 49 | 72 | 27 | 1 | 171 | 29 | 0.145 | 0.373 |
| Aarabi[23] | 2010 | 54/99 | 21 | 23 | 10 | 65 | 43 | 31 | 66 | 2 | 128 | 70 | 0.353 | <0.001 |
| Idali[12] | 2012 | 106/100 | 35 | 53 | 18 | 123 | 89 | 72 | 27 | 1 | 171 | 29 | 0.145 | 0.373 |
| Khosravi[24] | 2013 | 595/100 | 128 | 208 | 85 | 464 | 378 | 72 | 27 | 1 | 171 | 29 | 0.145 | 0.373 |
| Shakarami[25] | 2015 | 100/100 | 33 | 50 | 17 | 116 | 84 | 45 | 50 | 5 | 140 | 60 | 0.300 | 0.056 |
MAFs, minor allele frequencies; HWE, Hardy-Weinberg equilibrium
Quantitative synthesis
Table 3 listed the main results of the meta-analysis of MTHFR C677T and A1298C polymorphisms and RPL risk in the Iranian population. When all the eligible studies were pooled into the meta-analysis of MTHFR C677T polymorphism, a significant increased risk of RPL was observed in the allelic model (T vs. C: OR = 1.700, 95% CI = 1.208-2.393, p = 0.002), the heterozygote model (CT vs. CC: OR = 1.670, 95% CI = 1.215-2.295, p = 0.002), the homozygote model (TT vs. CC: OR = 2.409, 95% CI = 1.291-4.497, p = 0.006), the dominant model (TT + CT vs. CC: OR = 1.847, 95% CI = 1.264-2.699, p = 0.002, Fig. 1A), and the recessive model (TT vs. CT + CC: OR = 1.858, 95% CI = 1.087-3.177, p = 0.024). In addition, when all the eligible studies were pooled into the meta-analysis of MTHFR A1298C polymorphism, a significant association was observed in the allelic model (C vs. A: OR = 3.190, 95% CI = 1.467-6.936, p = 0.003), the heterozygote model (AC vs. AA: OR = 0.344, 95% CI = 1.344-8.321, p = 0.009), the homozygote model (CC vs. AA: OR = 5.073, 95% CI = 1.710-15.051, p = 0.003, Fig. 1B), the dominant model (CC + AC vs. AA: OR = 4.006, 95% CI = 1.578-10.169, p = 0.003), and the recessive model (CC vs. AC + AA: OR = 5.061, 95% CI = 1.668-15.361, p = 0.004).
Table 3.
The meta-analysis of thrombophilic genes polymorphisms and RPL risk
| Polymorphism | Study Number | Genetic Model | Type of Model | Heterogeneity | Odds Ratio | Publication Bias | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| I2 (%) | PH | OR | 95% CI | Ztest | POR | PBeggs | PEggers | ||||
| MTHFR C677T | |||||||||||
| 11 | T vs. C | Random | 83.71 | <0.001 | 1.700 | 1.208-2.393 | 3.044 | 0.002 | 0.533 | 0.854 | |
| 11 | TC vs. CC | Random | 64.10 | 0.002 | 1.670 | 1.215-2.295 | 3.163 | 0.002 | 0.275 | 0.459 | |
| 10 | TT vs. CC | Random | 69.40 | 0.001 | 2.409 | 1.291-4.497 | 2.761 | 0.006 | 0.107 | 0.132 | |
| 11 | TT + TC vs. CC | Random | 78.55 | <0.001 | 1.847 | 1.264-2.699 | 3.171 | 0.002 | 0.640 | 0.743 | |
| 10 | TT vs. TC + CC | Random | 60.50 | 0.007 | 1.858 | 1.087-3.177 | 2.264 | 0.024 | 0.020 | 0.056 | |
| MTHFR A1298C | |||||||||||
| 9 | C vs. A | Random | 94.60 | <0.001 | 3.190 | 1.467-6.936 | 2.928 | 0.003 | 0.754 | 0.752 | |
| 9 | CA vs. AA | Random | 92.68 | <0.001 | 3.344 | 1.344-8.321 | 2.595 | 0.009 | 0.602 | 0.995 | |
| 9 | CC vs. AA | Random | 70.70 | <0.001 | 5.073 | 1.710-15.051 | 2.927 | 0.003 | 0.076 | 0.015 | |
| 9 | CC + CA vs. AA | Random | 93.67 | <0.001 | 4.006 | 1.578-10.169 | 2.920 | 0.003 | 0.916 | 0.937 | |
| 9 | CC vs. CA + AA | Random | 74.31 | <0.001 | 5.061 | 1.668-15.361 | 2.863 | 0.004 | 0.348 | 0.022 | |
| Prothrombin G20210A | |||||||||||
| 4 | A vs. G | Fixed | 45.61 | 0.138 | 1.979 | 1.128-3.472 | 2.379 | 0.017 | 0.308 | 0.902 | |
| 4 | AA + AG vs. GG | Fixed | 52.94 | 0.095 | 2.060 | 1.162-3.652 | 2.474 | 0.013 | 0.308 | 0.881 | |
| FVL 1619 G/A | |||||||||||
| 6 | A vs. G | Fixed | 41.08 | 0.131 | 2.252 | 1.504-3.373 | 3.942 | <0.001 | 0.452 | 0.572 | |
| 5 | AG vs. GG | Fixed | 43.13 | 0.134 | 1.695 | 0.980-2.932 | 1.888 | 0.059 | 0.462 | 0.786 | |
| 3 | AA vs. GG | Fixed | 0.00 | 0.995 | 3.277 | 0.861-12.473 | 1.740 | 0.082 | 1.000 | 0.425 | |
| 6 | AA + AG vs. GG | Fixed | 44.02 | 0.112 | 2.217 | 1.447-3.395 | 3.658 | <0.001 | 0.452 | 0.626 | |
| 3 | AA vs. AG + GG | Fixed | 0.00 | 0.985 | 2.867 | 0.756-10.868 | 1.549 | 0.121 | 1.000 | 0.333 | |
| PAI-1 4G/5G | |||||||||||
| 5 | 4G vs. 5G | Random | 75.26 | 0.003 | 2.159 | 1.427-3.244 | 3.244 | <0.001 | 0.806 | 0.925 | |
| 5 | 4G/5G vs. 5G/5G | Random | 86.95 | <0.001 | 1.799 | 0.852-3.796 | 1.541 | 0.123 | 0.220 | 0.087 | |
| 5 | 4G/4G vs. 5G/5G | Fixed | 32.94 | 0.202 | 9.811 | 4.782-20.130 | 6.227 | <0.001 | 0.462 | 0.071 | |
| 5 | 4G/4G + 4G/5G vs. 5G/5G | Random | 79.84 | 0.001 | 1.961 | 1.104-3.484 | 2.298 | 0.022 | 0.806 | 0.427 | |
| 5 | 4G/4G vs. 4G/5G + 5G/5G | Fixed | 22.62 | 0.270 | 10.161 | 4.975-20.754 | 6.363 | <0.001 | 0.462 | 0.050 | |
Fig. 1.
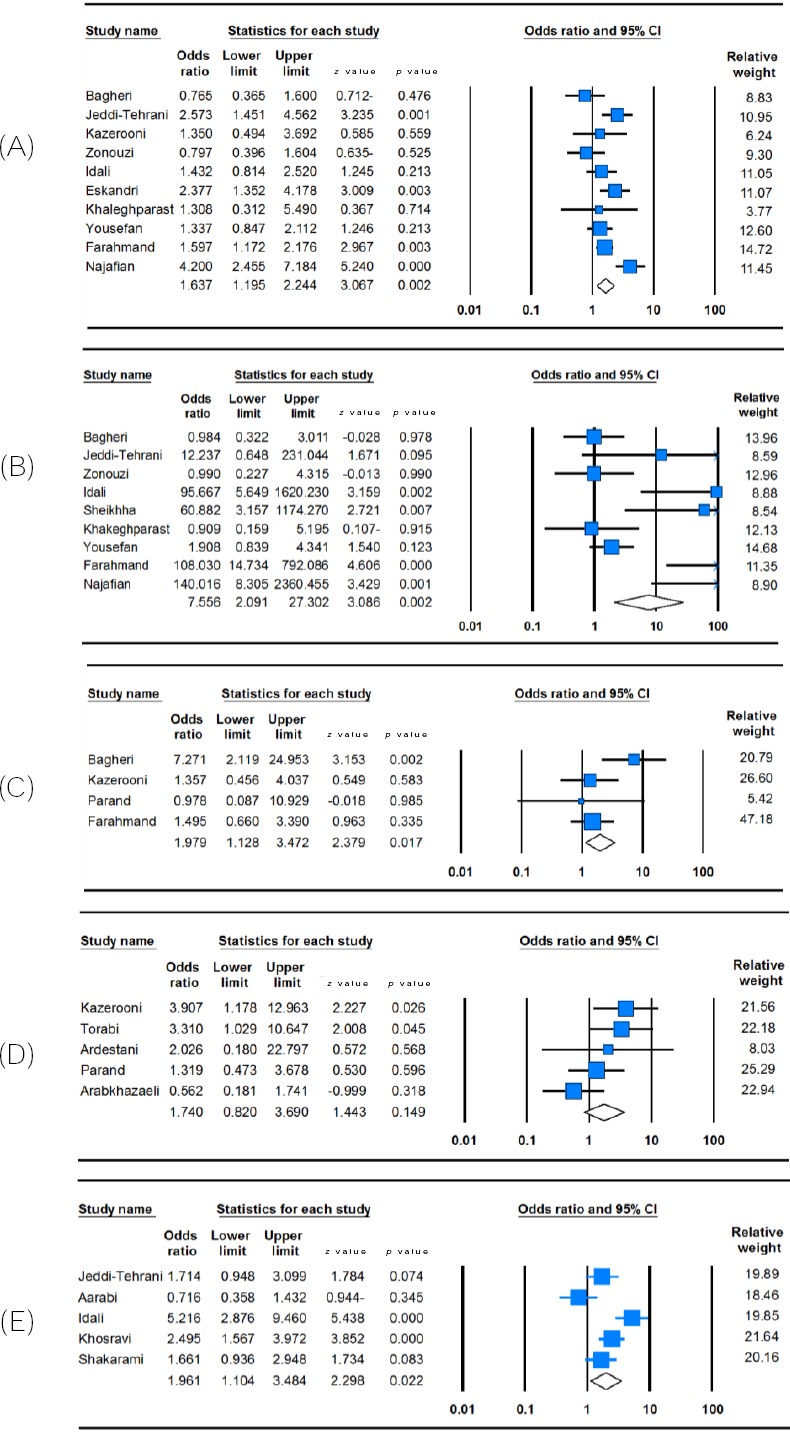
Forest plot of RPL susceptibility associated with thrombophilic genes polymorphisms. (A) MTHFR C677T (dominant model: TT + CT vs. CC); (B) MTHFR A1298C (homozygote model; AA vs. CC); (C) Prothrombin G20210A (allele model; A vs. G); (D) FVL G1619A (heterozygote model: GA vs. GG), and (E) PAI-1 4G/5G (dominant model: 4G/4G + 4G/5G vs. 5G/5G). For each study, the estimation of OR and its 95% CI are plotted with a square and a horizontal line. A diamond indicates the pooled OR with 95% CI.
Table 3 summarizes the ORs with corresponding 95% CIs for association of Prothrombin G20210A, FVL G1691A, and PAI-1 4G/5G polymorphisms with RPL risk in the Iranian population. Among the eligible studies were pooled to the meta-analysis of Prothrombin G20210A polymorphism, only allelic and dominant model was applicable because they provided the genotypes of AA + GA vs. GG. Therefore, the pooled analyses showed a significant association between Prothrombin G20210A polymorphism and RPL in the Iranian population in the allelic model (A vs. G: OR = 1.979, 95% CI = 1.128-3.472, p = 0.017, Fig. 1C) and the dominant model (AA + GA vs. GG: OR = 2.060, 95% CI = 1.162-3.652, p = 0.013). When all the eligible studies were pooled into the meta-analysis of FVL G1691A polymorphism, we observed a significant increased risk of RPL under allelic model (A vs. G: OR = 2.252, 95% CI = 1.504-3.373, p < 0.001, Fig. 1D) and dominant model (AA+GA vs. GG: OR = 2.217, 95% CI = 1.447-3.395, p < 0.001), but not in the heterozygote model (AG vs. GG: OR = 1.695, 95% CI = 0.980-2.932, p = 0.059), the homozygote (AA vs. GG: OR = 3.277, 95% CI = 0.861-12.473, p = 0.082), and the recessive model (AA vs. AG + GG: OR=2.867, 95% CI = 0.756-10.868, p = 0.121). In addition, there was a significant association between PAI-1 4G/5G polymorphism and RPL in the
Iranian population in the allelic model (4G vs. 5G: OR = 2.159, 95% CI = 1.427-3.244, p < 0.001), the homozygote model (4G/4G vs. 5G/5G: OR = 9.811, 95% CI = 4.782-20.130, p < 0.001), the dominant model (4G/4G + 4G/5G vs. 5G/5G: OR = 1.961, 95% CI = 1.104-3.484, p = 0.022, Fig. 1E), and the recessive model (4G/4G vs. 4G/5G + 5G/5G: OR = 10.161, 95% CI = 4.975-20.754, p < 0.001), but not in the heterozygote model (4G/5G vs. 5G/5G: OR = 1.799, 95% CI = 0.852-3.796, p = 0.123).
Sensitivity analysis
To evaluate the influence of individual studies on the risk of RPL, the studies were sequentially deleted from this meta-analysis, and the pooled ORs were performed. However, the results did not change exactly, which verify that no individual studies significantly affected the pooled ORs. Additionally, sensitivity analysis was performed after excluding HWE-violating studies, and the corresponding pooled ORs were not materially altered, indicating that our results are statistically robust (not shown).
Publication Bias
In this meta-analysis, Begg’s funnel plot and Egger’s test were used to assess the publication bias of included studies. The funnel plot revealed no obvious publication bias for MTHFR A1298C, Prothrombin G20210A, FVL G1691A, and PAI-1 4G/5G, and this result was confirmed by Begg’s test and Egger’s test (Fig. 2). However, the shapes of the funnel plots revealed obvious asymmetry for MTHFR C677T and MTHFR A1298C in the recessive model (Fig. 2A), suggesting that there were obvious publication biases in these two genetic models. Moreover, the results of Egger’s regression test provided sufficient evidence for publication bias for MTHFR A1298C in the recessive model (PBegg’s = 0.348, PEgger’s = 0.022), but not for MTHFR C677T (PBegg’s = 0.020, PEgger’s = 0.056). Therefore, we used the Duval and Tweedie non-parametric “trim-and-fill” method to adjust the results of publication bias for MTHFR A1298C polymorphism recessive model. However, the meta-analysis with and without ‘‘trim-and-fill’’ method did not show different results, showing that the results of this meta-analysis are statistically robust.
Fig. 2.
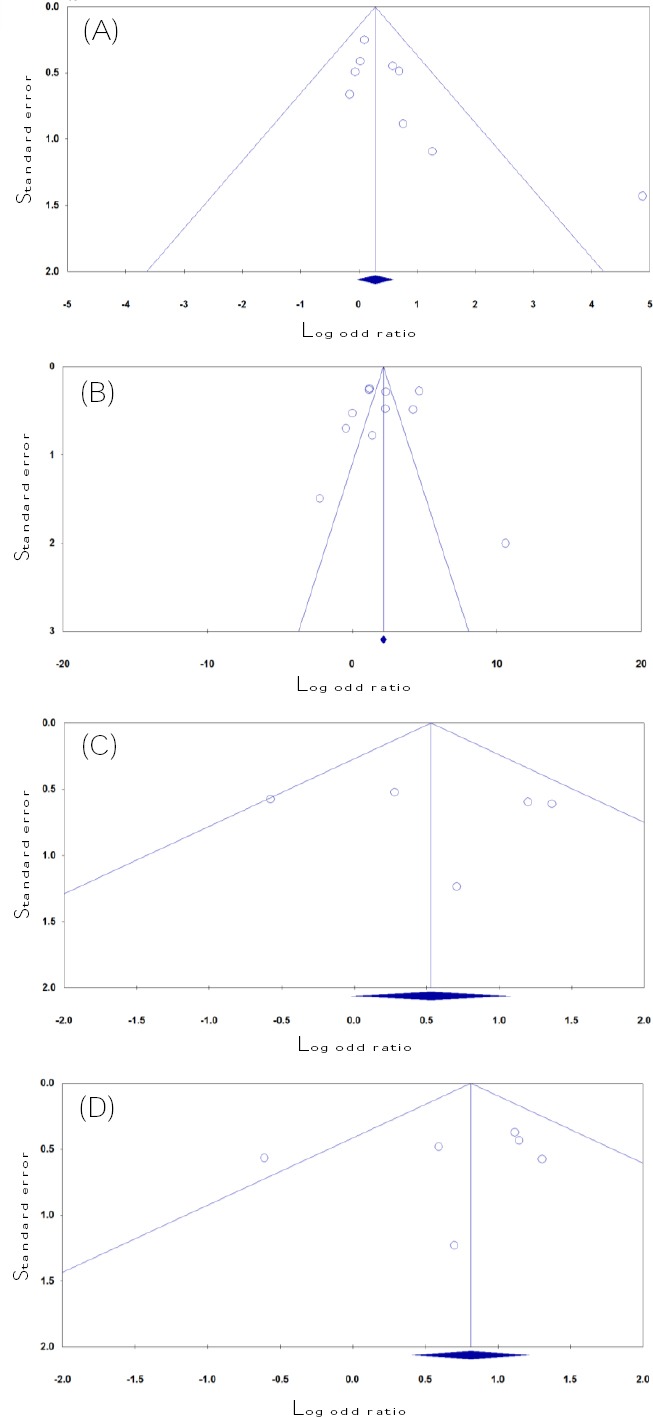
Begg’s funnel plots for thrombophilia gene polymorphisms and RPL risk in the Iranian patients to test the publication bias. (A) MTHFR C677T (recessive model: TT vs. CT + CC); (B) MTHFR A1298C (dominant model: AA vs. AC + CC); (C) Prothrombin G20210A (dominant model: AA + GA vs. GG); (D) FVL G1691A (allele model: A vs. G). Each point represents a separate study for the indicated association.
DISCUSSION
It is known that folate is required for the proper development of fetus and placenta[32]. Aberrations in folate pathway such as maternal folate deficiency, maternal hyperhomocysteinemia, and either MTHFR C677T or A1298C polymorphisms were found to contribute to the etiology of RPL in different populations[33]. Although MTHFR A1298C poly-morphism may not responsible for increased total homocysteine, it seems that this polymorphism contributes significantly to the increased homocysteine levels[34,35]. Previous studies have reported that MTHFR C677T polymorphism is significantly associated with the increased risk of RPL. For instance, Chen et al.[36] in a meta-analysis of 16 articles involving 1420 cases with RPL and 1408 controls reported that MTHFR C677T was significantly associated with RPL risk in the Chinese population under all genetics models. Similarly, Wu et al.[37] and Cao et al.[38] findings supported that the idea that MTHFR C677T polymorphism was associated with the increased risk of RPL among Asians, but not Caucasians. Based on these studies, the MTHFR 677TT polymorphism has a significant increased likelihood of RPL in Asians, which is in agreement with our conclusion. The data of a meta-analysis by Nair et al.[39] showed that MTHFR A1298C polymorphism was a genetic risk factor for RPL. However, Cao et al.[38] did not find any significant association between MTHFR A1298C polymorphism and RPL susceptibility. The combined data, based on previous studies, showed that both MTHFR C677T and MTHFR A1298C polymorphisms might be a risk factor for RPL.
As for the other two polymorphisms, we also find a significant association of polymorphisms in Prothrombin G20210A, FVL G1691A, and PAI-1 4G/5G with the risk of RPL, which were consistent with the majority but not all previous studies[40-42]. In a meta-analysis conducted by Gao et al.[41], the Prothrombin G20210A variant was reported to increase the risk of RPL (fetal loss, primary RPL, or secondary RPL)[43], particularly in Europeans and women older than 29 years. In another meta-analysis, Kovalevsky et al.[44] found that FVL G1691A or Prothrombin gene polymorphisms were associated with the increased risk of two or more miscarriages compared with women without these polymorphisms. In addition, in three meta-analyses, Chen et al.[45], Su et al.[46] and Li et al.[47] suggested that PAI-1 4G/5G polymorphism might be associated with RPL development.
Between-study heterogeneity, a multifactorial phoneme in meta-analyses, is a potential problem when interpreting the results[48-50]. In addition to ethnicity and the source of controls, selection of controls, race variation, age, gender, and prevalence of lifestyle factors might also generate the heterogeneity[35,48,49]. In the present meta-analysis, between-study heterogeneity was observed in all polymorphisms, and thus a random-effect model was used for those genetic models.
There was an inevitable publication bias in our meta-analysis because we retrieved only published studies. Publication bias was assessed by funnel plots whose symmetries were further evaluated by Egger’s linear regression tests. We have suggested that for the recessive model, the publication bias might be owing to the limited number of the selected studies. Therefore, the negative results in our meta-analysis are possibly due to the limited number of publications to determine statistical significance.
To the best of our knowledge, there is no earlier study on the analysis of thrombophilia gene polymorphisms in RPL in the Iranian population. However, in interpreting results of this meta-analysis, some limitations should be addressed. First, there was no sufficient number of relevant studies to explore more comprehensive association between the thrombophilic genes and RPL in the Iranian patients. Second, in this meta-analysis, only published studies were searched. It is possible that some important unpublished studies that meet our inclusion criteria were missed and ignored in the literature search. Therefore, inevitable publication bias might be exist, which could eventually help explain the possible existence of publication bias in the recessive model. Third, the Iranian population is mixed of different ethnicities, including Persian, Azeri, Kurdish, Lurs, Gilaki, Balochi, etc. However, we did not conduct subgroup analyses because insufficient data were available from the primary literature search. Moreover, due to limited individual data, a more precise analysis on other covariates, such as age, number of abortions, and environmental factors should be performed. As a result, more studies with large sample sizes in view of these factors are also desired. Finally, due to the lack of the original data, we did not take potential interactions among gene-gene (especially thrombo-philic genes interactions), gene-environment, or even different polymorphism loci of the same gene, which all may affect RPL risk in the population.
In conclusion, based on the available evidence, the current meta-analysis demonstrates that there is a significant association between the MTHFR C677T, MTHFR A1298C, Prothrombin G20210A, FVL G1691A, and PAI-1 4G/5G polymorphisms and RPL risk in the Iranian population. In addition, the direction of further research with a larger sample size should focus not only on the simple relationship of thrombophilic genes polymorphisms and RPL risk but also on gene-gene and gene-environment interactions.
Footnotes
CONFLICT OF INTEREST. None declared.
REFERENCES
- 1.McPherson E. Recurrence of stillbirth and second trimester pregnancy loss. American journal of medical genetics. 2016;170A(5):1174–1180. doi: 10.1002/ajmg.a.37606. [DOI] [PubMed] [Google Scholar]
- 2.Kumar N, Ahluwalia J, Das R, Rohilla M, Bose S, Kishan H, Varma N. Inherited thrombophilia profile in patients with recurrent miscarriages:Experience from a tertiary care center in north India. Obstetrics and gynecology science journal. 2015;58(6):514–517. doi: 10.5468/ogs.2015.58.6.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diejomaoh MF. recurrent spontaneous miscarriage is still a challenging diagnostic and therapeutic quagmire. Medical principles and practice. 2015;24(Suppl 1):38–55. doi: 10.1159/000365973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris EN, Chan JK, Asherson RA, Aber VR, Gharavi AE, Hughes GR. Thrombosis, recurrent fetal loss, and thrombocytopenia. Archives of internal medicine journal. 1986;146(11):2153–2156. [PubMed] [Google Scholar]
- 5.Neamatzadeh H, Ramazani V, Kalantar SM, Ebrahimi M, Sheikhha MH. Serum immune reactivity against β2-Glycoprotein-I and anti-neutrophil cytoplasmic auto-antibodies by ELI-P-Complex screening technology in recurrent miscarriage. Minerva ginecologica. 2016;68(3):243–249. [PubMed] [Google Scholar]
- 6.Aytekin E, Ergun S, Ergun M, Percin F. Evaluation of geno flow thrombophilia array test kit in its detection of mutations in Factor V Leiden (G1691A), prothrombin G20210A, MTHFR C677T and A1298C in blood samples from 113 Turkish female patients. Genetic testing and molecular biomarkers. 2014;18(11):717–721. doi: 10.1089/gtmb.2014.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.López-Jiménez J, Porras-Dorantes Á, Juárez-Vázquez C, García-Ortiz J, Fuentes-Chávez C, Lara-Navarro I, Jaloma-Cruz Molecular thrombophilic profile in Mexican patients with idiopathic recurrent pregnancy loss. Genetics and molecular research. 2016;15(4) doi: 10.4238/gmr.15048728. doi:10.4238/gmr.15048728. [DOI] [PubMed] [Google Scholar]
- 8.Bagheri M, Abdi Rad I, Omrani MD, Nanbakhsh F. C677T and A1298C mutations in the Methylene-tetrahydrofolate reductase gene in patients with recurrent abortion from the Iranian Azeri Turkish. International journal of fertility and sterility. 2010;4(3):134–139. [Google Scholar]
- 9.Jeddi-Tehrani M, Torabi R, Zarnani AH, Mohammadzadeh A, Arefi S, Zeraati H, Akhondi MM, Chamani-Tabriz L, Idali F, Emami S, Zarei S. Analysis of plasminogen activator inhibitor-1, integrin beta3, beta fibrinogen, and methylenetetrahydrofolate reductase polymorphisms in Iranian women with recurrent pregnancy loss. The American journal of reproductive immunolog. 2011;66(2):149–156. doi: 10.1111/j.1600-0897.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- 10.Kazerooni T, Ghaffarpasand F, Asadi N, Dehkhoda Z, Dehghankhalili M, Kazerooni Y. Correlation between thrombophilia and recurrent pregnancy loss in patients with polycystic ovary syndrome:A comparative study. The Chinese medical association. 2013;76:282–288. doi: 10.1016/j.jcma.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Poursadegh Zonouzi A, Chaparzadeh N, Asghari Estiar M, Mehrzad Sadaghiani M, Farzadi L, Ghasemzadeh A, Sakhinia M, Sakhinia E. Methylenetetrahydrofolate reductase C677T and A1298C mutations in women with recurrent spontaneous abortions in the Northwest of Iran. ISRN Obstetric and gynecology. 2012;2012:945486. doi: 10.5402/2012/945486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Idali F, Zareii S, Mohammad-Zadeh A, Reihany-Sabet F, Akbarzadeh-Pasha Z, Khorram-Khorshid H, Zarnani AH, Jeddi-Tehrani M. Plasminogen activator inhibitor 1 and methylenetetrahydrofolate reductase gene mutations in iranian women with polycystic ovary syndrome. American journal of reproductive immunology. 2012;68:400–407. doi: 10.1111/aji.12002. [DOI] [PubMed] [Google Scholar]
- 13.Eskandari F, Akbari MT, Zare Karizi S. Association of C677T and A1298C polymorphisms of the MTHFR gene with recurrent pregnancy loss. Pajoohandeh journal. 2013;18:167–173. [Google Scholar]
- 14.Khaleghparast A, Khaleghparast S, Khaleghparast H. Association between the A1298C polymorphism of the methylenetetrahydrofolate reductase gene and recurrent spontaneous abortion. Iranian journal of neonatology. 2014;5(2):7–11. [Google Scholar]
- 15.Yousefian E, Taghi Kardi M, Allahveisi A. Methylenetetrahydrofolate reductase C677T and A1298C polymorphism in Iranian women with idiopathic recurrent pregnancy losses. Iranian Red Crescent medical journal. 2014;16(7):e16763. doi: 10.5812/ircmj.16763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farahmand K, Totonchi M, Hashemi M, Reyhani Sabet F, Kalantari H, Gourabi H, Mohseni Meybodi A. Thrombophilic genes alterations as risk factor for recurrent pregnancy loss. The journal of maternal-fetal and neonatal medicine. 2015;29(8):1269–1273. doi: 10.3109/14767058.2015.1044431. [DOI] [PubMed] [Google Scholar]
- 17.Najafian M, Yar Ahmadi E, Mohammadi Asl J, Shariati G, Yar Ahmadi N. Study the correlation between polymorphism of MTHFR thrombophilic genes and pregnancy loss in Ahvaz city. Biosciences biotechnology research asia. 2016;13(2):681–686. [Google Scholar]
- 18.Sheikhha MH, Kalantar SM, Ghasemi N, Soleimanian S. Association between MTHFR 1298A>C polymorphism with RSA and IVF Failure. Iranian journal of pediatric hematology and oncology. 2012;2(3):109–115. [Google Scholar]
- 19.Arabkhazaeli N, Ghanaat K, Hashemi-Soteh MB. H1299R in coagulation factor V and Glu429Ala in MTHFR genes in recurrent pregnancy loss in Sari, Mazandaran. International journal of reproductive biomedicine. 2016;14(5):329–334. [PMC free article] [PubMed] [Google Scholar]
- 20.Teremmahi Ardestani M, Nodushan HH, Aflatoonian A, Ghasemi N, Sheikhha MH. Case control study of the Factor V Leiden and factor II G20210A mutation frequency in women with recurrent miscarriage loss. International journal of reproductive biomedicine. 2013;11(1):61–64. [PMC free article] [PubMed] [Google Scholar]
- 21.Parand A, Zolghadri J, Nezam M, Afrasiabi A, Haghpanah S, Karimi M. Inherited thrombophilia and recurrent pregnancy loss. Iranian Red Crescent medical journal. 2013;15(12):e13708. doi: 10.5812/ircmj.13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torabi R, Zarei S, Zeraati H, Zarnani AH, Akhondi MM, Hadavi R, Shiraz ES, Jeddi-Tehrani M. Combination of thrombophilic gene polymorphisms as a cause of increased the risk of recurrentpregnancy loss. Journal of reproduction and infertility journal. 2012;13(2):89–94. [PMC free article] [PubMed] [Google Scholar]
- 23.Aarabi M, Memariani T, Arefi S, Aarabi M, Hantoosh Zadeh S, Akhondi MA, Modarressi MH. Polymorphisms of plasminogen activator inhibitor-1, angiotensin converting enzyme and coagulation factor XIII genes in patients with recurrent spontaneous abortion. The journal of maternal-fetal and neonatal medicine. 2010;24(3):545–548. doi: 10.3109/14767058.2010.511331. [DOI] [PubMed] [Google Scholar]
- 24.Khosravi F, Zarei S, Ahmadvand N, Akbarzadeh-Pasha Z, Savadi E, Zarnani A, Sadeghi MR, Jeddi-Tehrani M. Association between plasminogen activator inhibitor 1 gene mutation and different subgroups of recurrent miscarriage and implantation failure. The journal of assisted reproduction and genetics. 2013;31(1):121–124. doi: 10.1007/s10815-013-0125-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shakarami F, Akbari MT, Zare Karizi S. Association of plasminogen activator inhibitor-1 and angiotensin converting enzyme polymorphisms with recurrent pregnancy loss in Iranian women. International journal of reproductive biomedicine. 2015;13(10):627–632. [PMC free article] [PubMed] [Google Scholar]
- 26.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DerSimonian R, Laird N. Meta-analysis in clinical trials. Controlled clinical trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 28.Salanti G, Amountza G, Ntzani E, Ioannidis J. Hardy–Weinberg equilibrium in genetic association studies:an empirical evaluation of reporting, deviations, and power. The European journal of human genetics. 2005;13(7):840–848. doi: 10.1038/sj.ejhg.5201410. [DOI] [PubMed] [Google Scholar]
- 29.Thabane L, Mbuagbaw L, Zhang S, Samaan Z, Marcucci M, Ye C, Thabane M, Giangregorio L, Dennis B, Kosa D, Borg Debono V, Dillenburg R, Fruci V, Bawor M, Lee J, Wells G, Goldsmith CH. A tutorial on sensitivity analyses in clinical trials:the what, why, when and how. BMC medical research methodology. 2013;13:92. doi: 10.1186/1471-2288-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 31.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fekete K, Berti C, Trovato M, Lohner S, Dullemeijer C, Souverein O, Cetin I, Decsi T. Effect of folate intake on health outcomes in pregnancy:a systematic review and meta-analysis on birth weight, placental weight and length of gestation. Nutrition journal. 2012;11:75. doi: 10.1186/1475-2891-11-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebadifar A, KhorramKhorshid HR, Kamali K, Salehi Zeinabadi M, Khoshbakht T, Ameli N. Maternal supplementary folate intake, methylenetetrahy-drofolate reductase (mthfr) c677t and a1298c polymorphisms and the risk of orofacial cleft in iranian children. The Avicenna journal of medical biotechnology. 2015;7(2):80–84. [PMC free article] [PubMed] [Google Scholar]
- 34.Klai S, Fekih-Mrissa N, El Housaini S, Kaabechi N, Nsiri B, Rachdi R, Gritli N. association of MTHFR A1298C polymorphism (but not of MTHFR C677T) with elevated homocysteine levels and placental vasculopathies. Blood coagulation and fibrinolysis. 2011;22(5):374–378. doi: 10.1097/MBC.0b013e328344f80f. [DOI] [PubMed] [Google Scholar]
- 35.Abedinzadeh M, Zare-Shehneh M, Neamatzadeh H, Abedinzadeh M, Karami H. Association between MTHFR C677T polymorphism and risk of prostate cancer:evidence from 22 studies with 10,832 cases and 11,993 controls. Asian Pacific journal of cancer prevention. 2015;16(11):4525–4530. doi: 10.7314/apjcp.2015.16.11.4525. [DOI] [PubMed] [Google Scholar]
- 36.Chen H, Yang X, Lu M. Methylenetetrahydrofolate reductase gene poly-morphisms and recurrent pregnancy loss in China:a systematic review and meta-analysis. Archives of gynecology and obstetrics. 2016;293(2):283–290. doi: 10.1007/s00404-015-3894-8. [DOI] [PubMed] [Google Scholar]
- 37.Wu X, Zhao L, Zhu H, He D, Tang W, Luo Y. Association between the MTHFR C677T polymorphism and recurrent pregnancy loss:a meta-analysis. Genetic testing and molecular biomarkers. 2012;16(7):806–811. doi: 10.1089/gtmb.2011.0318. [DOI] [PubMed] [Google Scholar]
- 38.Cao Y, Xu J, Zhang Z, Huang X, Zhang A, Wang J, Zheng Q, Fu L, Du J. Association study between methylenetetrahydrofolate reductase polymorphisms and unexplained recurrent pregnancy loss:A meta-analysis. Gene. 2013;514:105–111. doi: 10.1016/j.gene.2012.10.091. [DOI] [PubMed] [Google Scholar]
- 39.Nair R, Khanna A, Singh R, Singh K. Association of maternal and fetal MTHFR A1298C polymorphism with the risk of pregnancy loss:a study of an Indian population and a meta-analysis. Fertility and sterility. 2013;99(5):1311–1318. doi: 10.1016/j.fertnstert.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 40.Rai V. Methylenetetrahydrofolate reductase gene A1298C polymorphism and susceptibility to recurrent pregnancy loss:a meta-analysis. Cellular and molecular biology. 2014;60(2):27–34. [PubMed] [Google Scholar]
- 41.Gao H, Tao FB. Prothrombin G20210A mutation is associated with recurrent pregnancy loss:A systematic review and meta-analysis update. Thrombosis research. 2015;135(2):339–346. doi: 10.1016/j.thromres.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Sergi C, Al Jishi T, Walker M. Factor V Leiden mutation in women with early recurrent pregnancy loss:a meta-analysis and systematic review of the causal association. Archives of gynecology and obstetrics. 2014;291(3):671–679. doi: 10.1007/s00404-014-3443-x. [DOI] [PubMed] [Google Scholar]
- 43.Li X, Liu Y, Zhang R, Tan J, Chen L, Liu Y. Meta-analysis of the association between plasminogen activator inhibitor-1 4G/5G polymorphism and recurrent pregnancy loss. Medical science monitor. 2015;21:1051–1056. doi: 10.12659/MSM.892898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kovalevsky G, Gracia CR, Berlin JA, Sammel MD, Barnhart KT. Evaluation of the association between hereditary thrombophilias and recurrent pregnancy loss. Archives of internal medicine. 2004;164(5):558–563. doi: 10.1001/archinte.164.5.558. [DOI] [PubMed] [Google Scholar]
- 45.Chen H, Nie S, Lu M. Association between Plasminogen Activator Inhibitor-1 Gene polymorphisms and recurrent pregnancy loss:a systematic review and meta-analysis. The American journal of reproductive immunology. 2014;73(4):292–300. doi: 10.1111/aji.12321. [DOI] [PubMed] [Google Scholar]
- 46.Su M, Lin S, Chen Y, Kuo P. Genetic association studies of ACE and PAI-1 genes in women with recurrent pregnancy loss. Thrombosis and hemostasis. 2012;109(1):8–15. doi: 10.1160/TH12-08-0584. [DOI] [PubMed] [Google Scholar]
- 47.Li X, Liu Y, Zhang R, Tan J, Chen L, Liu Y. Meta-analysis of the association between plasminogen activator inhibitor-1 4G/5G polymorphism and recurrent pregnancy loss. Medical science monitor. 2015;21:1051–1056. doi: 10.12659/MSM.892898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sobhan MR, Forat Yazdi M, Mazaheri M, Zare Shehneh M, Neamatzadeh H. Association between the DNA repair gene XRCC3 rs↧39 polymorphism and risk of osteosarcoma:a systematic review and meta-analysis. Asian Pacific journal of cancer prevention. 2017;18(2):549–555. doi: 10.22034/APJCP.2017.18.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mehdinejad M, Sobhan MR, Mazaheri M, Zare Shehneh M, Neamatzadeh H, Kalantar SM. Genetic association between ERCC2, NBN, RAD51 gene variants and osteosarcoma risk:a systematic review and meta-analysis. Asian Pacific journal of cancer prevention. 2017;18(5):1315–1321. doi: 10.22034/APJCP.2017.18.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Namazi A, Abedinzadeh M, Nourbaksh P, Nematzadeh H. Association between the XRCC3 Thr241Met polymorphism and risk of colorectal cancer:a meta-analysis of 5,193 cases and 6,645 controls. Asian Pacific journal of cancer prevention. 2015;16(6):2263–2268. doi: 10.7314/apjcp.2015.16.6.2263. [DOI] [PubMed] [Google Scholar]


