ABSTRACT
The FDA-CDC Antimicrobial Resistance Isolate Bank was created in July 2015 as a publicly available resource to combat antimicrobial resistance. It is a curated repository of bacterial isolates with an assortment of clinically important resistance mechanisms that have been phenotypically and genotypically characterized. In the first 2 years of operation, the bank offered 14 panels comprising 496 unique isolates and had filled 486 orders from 394 institutions throughout the United States. New panels are being added.
KEYWORDS: AR Bank, AR Isolate Bank, antimicrobial resistance, antibiotic resistance
TEXT
Antimicrobial resistance is a cause for global concern. Antimicrobials not only treat primary bacterial infections but also make possible other medical advances, such as surgery, organ transplantation, and chemotherapy for cancer patients (1). The Centers for Disease Control and Prevention (CDC) has estimated that at least 2 million people become infected with antimicrobial-resistant bacteria every year and that at least 23,000 people die as a direct result of these infections (2). This problem has led to numerous “calls to action” from professional organizations, public health agencies, and governments, including the Infectious Diseases Society of America (IDSA), the American Society for Microbiology (ASM), the CDC, the United Kingdom and United States governments, the World Health Organization (WHO), and the United Nations (2–7).
In the United States, the urgent need to address antimicrobial resistance led to the issuance of Presidential Order 13676 on 23 September 2014 (8). This order identified critical actions to be taken by key federal departments and agencies to combat the rise of resistant bacteria. This action was followed by the development of the National Action Plan for Combating Antibiotic-Resistant Bacteria (CARB) in March 2015 (9). The goals of this plan are to (i) slow the emergence of resistant bacteria and prevent the spread of resistant infections, (ii) strengthen national one-health surveillance efforts to combat resistance, (iii) advance the development and use of rapid and innovative diagnostic tests for the identification and characterization of resistant bacteria, (iv) accelerate basic and applied research and the development of new antibiotics, therapeutics, and vaccines, and (v) improve international collaboration and capacities for antibiotic resistance prevention, surveillance, control, and antibiotic research and development. An objective under CARB goal number 2 was to establish an isolate bank of resistant bacterial strains to facilitate development and evaluation of diagnostic tests and treatments. This resource would help diagnostic and pharmaceutical companies faced with the challenge of having limited access to isolates to develop rapid, innovative diagnostic tests and new antimicrobial agents.
To accomplish these goals, an interagency agreement was initiated between the Center for Devices and Radiological Health (CDRH) at the U.S. Food and Drug Administration (FDA) and the National Center for Emerging and Zoonotic Infectious Diseases (NCEZID) at the CDC to develop a repository of bacterial isolates known as the FDA-CDC Antimicrobial Resistance Bank (AR Isolate Bank) (the name will change to the CDC-FDA Antibiotic Resistance Isolate Bank on 1 January 2018). The AR Isolate Bank can be found at https://www.cdc.gov/drugresistance/resistance-bank/index.html. A joint team of scientists from the FDA and CDC is responsible for overseeing the project, including data updates and additions to the AR Isolate Bank. All technical and administrative aspects, maintenance, and requests for isolates are handled by the CDC. The goal of the AR Isolate Bank is to serve as a collection of highly characterized isolates that are accessible to government agencies, academic institutions, clinical and commercial laboratories, public health institutions, pharmaceutical companies, and biotech and diagnostic companies developing in vitro diagnostic devices.
The AR Isolate Bank was launched in June 2015, and since then, isolates in the bank have been used for a variety of purposes. Public health, clinical, and reference laboratories use isolates as a resource for the verification and validation of new diagnostic tests and for implementation of updated breakpoints approved by the Clinical and Laboratory Standards Institute (CLSI). In vitro diagnostic-device manufacturers use isolates in studies to satisfy some of the regulatory requirements established by the CDRH at the FDA for devices that detect phenotypic susceptibility or molecular markers of resistance (10, 11). Pharmaceutical companies use isolates to test the microbiology profiles of antimicrobial agents in development, as outlined by the Center for Drug Evaluation and Research at the FDA (12). Finally, academic researchers use the isolates to understand more about known and novel mechanisms of resistance in order to design innovative diagnostic methods and therapeutics.
ORGANISMS IN THE AR ISOLATE BANK
Organisms added to the AR Isolate Bank are obtained mostly from CDC surveillance programs, outbreak investigations, and isolates sent to the CDC for reference testing. The CDC has also reached out to domestic and international partners to obtain isolates with new or unusual resistance that are not part of the existing CDC collection. In addition, pharmaceutical companies have provided bacterial isolates from their collections for deposit into the AR Isolate Bank so that medical laboratories can validate and implement clinical breakpoints for new drugs. Offers of isolates or collections of isolates for deposition into the AR Isolate Bank are welcome and may be emailed to ARbank@cdc.gov.
All isolates are characterized before they are placed into the bank. Characterization includes checks for purity and identification by matrix-assisted laser desorption–ionization time of flight mass spectrometry (MALDI-TOF MS) using a Biotyper 3.1 MALDI-TOF system (Bruker Daltonics, Billerica, MA). Analysis by 16S rRNA gene sequencing is used if further identification is needed. The sequencing data are analyzed using Kraken (http://ccb.jhu.edu/software/kraken/) and BLAST against reference genomes available in GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (13). Reference broth microdilution is performed to obtain antibiotic susceptibility testing (AST) data in accordance with CLSI standards (14, 15). Whole-genome sequencing (WGS) is also performed to achieve better understanding of the genetic basis of resistance in each isolate. The majority of isolates have been sequenced at the CDC, although the National Institute of Allergy and Infectious Diseases from the National Institutes of Health (NIH), the Broad Institute, and the Institute for Genome Sciences (IGS) at the University of Maryland have also contributed to sequencing. The sequencing approach has included different sequencing platforms, such as the Illumina MiSeq, the Illumina HiSeq 2500 (San Diego, CA), and the PacBio RSII (Silicon Valley, CA) systems.
Once isolates are fully characterized, aliquots are prepared by subculture on nonselective media from the original frozen aliquot. Selective pressure is applied to isolates known to lose their plasmid-associated resistance genes. Isolates are then incubated overnight at 35°C, and new aliquots are prepared using cryopreservation media. Newly prepared aliquots are stored at −70°C. One aliquot for each isolate is kept as a reference for every lot prepared. Twenty percent of isolates undergo confirmatory AST for each lot produced.
ORDERING ISOLATES FROM THE AR ISOLATE BANK
A full listing of AR Bank isolates and associated data currently offered is available online at https://www.cdc.gov/drugresistance/resistance-bank/currently-available.html. On the website, requestors can access isolate-level MIC results and susceptibility interpretations, a list of all resistance mechanisms detected, and accession numbers and links to complete genotypic information (raw sequence data and assembled genome data) via the NCBI website (https://www.ncbi.nlm.nih.gov/sra/). Storage and propagation procedures for each panel are also posted.
Isolates may be requested using an electronic form on the website. This form solicits the requestor's contact information, institution type, and intended use of the isolates requested (including the requester's need for isolates to support a regulatory submission to the FDA). Each requestor must provide the signature of a biosafety official who can verify that their receiving institution can appropriately handle biosafety level 2 (BSL-2) organisms and accepts the risks of handling highly resistant bacteria. The CDC screens each order to ensure the legitimacy of the request and the institution. Once this vetting process is complete, the order is approved and may be packed and shipped. There is no cost for the panels, but the requestor is responsible for shipping charges. Thus, a courier account number is also required at the time of order.
The AR Isolate Bank routinely accepts orders only from laboratories in the United States. Requests to ship isolates internationally are considered only in collaboration with the government of the receiving country. These requests are outside the standard operating procedure of the AR Isolate Bank. If a potential recipient would like to pursue this option, please contact the AR Isolate Bank at ARbank@cdc.gov with the specific request and a governmental contact (an official at the ministry of health or a public health agency).
PANELS CURRENTLY AVAILABLE IN THE AR ISOLATE BANK
The 2013 CDC AR Threats Report classified drug-resistant organisms into three priority categories: “urgent,” “serious,” and “concerning.” The FDA and CDC developed an initial list of medically important microorganisms, susceptibility profiles, and resistance mechanisms that comprised the top priorities set forth in the report (2). Using this list of priority isolates, the AR Isolate Bank was launched in June 2015.
Each panel in the AR Isolate Bank was developed with a particular use in mind, although other uses are also common. The first panel added was the Enterobacteriaceae carbapenem breakpoint panel (31 isolates), designed to help clinical laboratories implement and verify the new CLSI carbapenem breakpoints for Enterobacteriaceae. The second panel was the Gram-negative carbapenemase detection panel (80 isolates), designed to help laboratories verify tests for carbapenemase production (e.g., the modified Hodge test, the Carba NP test, or the modified carbapenem inactivation method test). The third panel was the Enterobacteriaceae carbapenemase diversity panel (53 isolates), which aimed to provide laboratories with a diversity of different carbapenemase types for many potential uses, including validation of molecular assays (13).
To enable detection of new resistance mechanisms and respond to evolving research and development needs, the collection of panels has expanded. As of June 2017, the AR Isolate Bank contained 14 panels comprising 496 unique isolates. Added panels include those focused on Neisseria gonorrhoeae (50 isolates), vancomycin-intermediate Staphylococcus aureus (14 isolates), Pseudomonas aeruginosa (44 isolates), Acinetobacter baumannii (41 isolates), and drug-resistant Candida spp. (other than C. albicans or C. auris) (32 isolates). The panel for isolates with new or novel antibiotic resistance was created as a flexible panel that houses isolates with unique or emerging resistance phenotypes or mechanisms (currently 9 isolates). This panel currently contains isolates with resistance to ceftazidime-avibactam and isolates that carry an mcr allele, conferring resistance to colistin. The ceftolozane-tazobactam panel (30 isolates) and ceftazidime-avibactam panel (30 isolates) were developed in collaboration with pharmaceutical companies to allow laboratories to verify AST of those drugs. The Candida auris panel (20 isolates) was added to allow investigators and laboratorians access to this emerging pathogen. The enteric pathogen diversity panel (30 isolates) was added to offer new genera (including Salmonella spp., Shigella spp., and Campylobacter spp.). The borderline oxacillin susceptibility panel (32 isolates) was added to provide isolates to challenge diagnostic methicillin-resistant Staphylococcus aureus testing.
IMPACT OF THE AR ISOLATE BANK
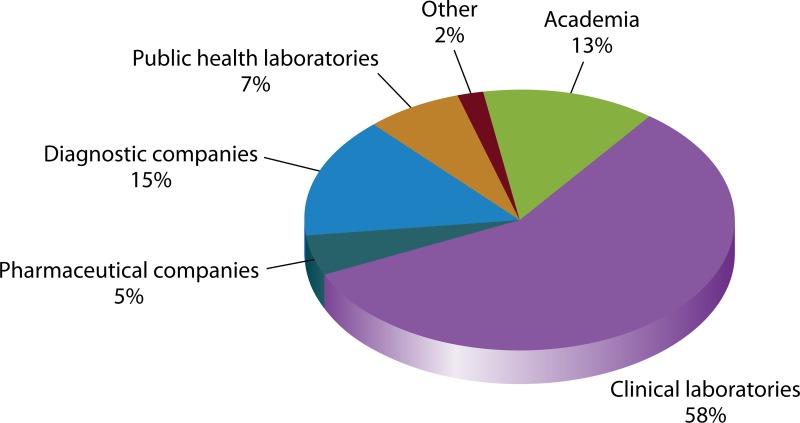
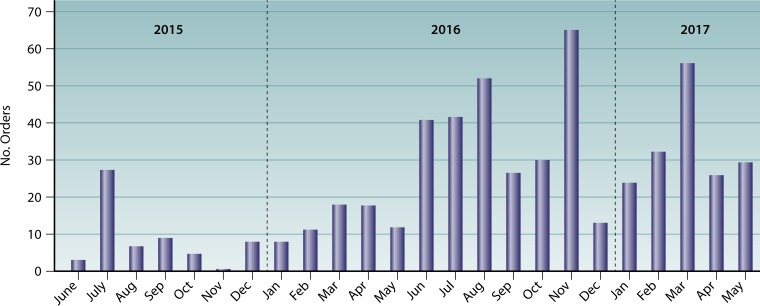
In its first 2 years, the AR Bank processed 486 orders and shipped over 44,000 aliquots to 394 institutions throughout the United States. These institutions include academic research laboratories, public health laboratories, academic medical centers, public and private hospitals, commercial laboratories, commercial pharmaceutical companies, commercial diagnostic companies, and several U.S. federal agencies (Fig. 1 and 2). Orders range widely in their intended use. Some panels provide challenge sets for the evaluation of diagnostic devices and new pharmaceutical agents and were used to provide data for FDA review. Organisms included in other panels have served as reference organisms for the verification and validation of new tests introduced into clinical laboratories and public health laboratories, including the CDC's Antibiotic Resistance Laboratory Network. Panels have also provided subject matter for advancing basic science research. To these ends, the most frequently ordered panels include the Gram-negative carbapenemase detection panel (223 orders), the Enterobacteriaceae carbapenemase diversity panel (204 orders), the Enterobacteriaceae carbapenem breakpoint panel (157 orders), and the Candida auris panel (115 orders).
FIG 1.
Panel requests by organization type. Clinical laboratories include academic medical centers, VA hospitals, military hospitals, private hospitals, public hospitals, private laboratories, and reference laboratories. “Academia” represents university research laboratories.
FIG 2.
Numbers of orders requested by month since the AR Isolate Bank's launch in June 2015. The bar graph represents the numbers of orders by month.
OTHER AR ISOLATE REPOSITORIES
For those looking for additional types of isolate collections, the following resources are available. The Walter Reed Army Institute of Research (WRAIR) maintains a collection of isolates through its Multidrug-Resistant Organism Repository and Surveillance Network (MRSN), available at http://www.wrair.army.mil/OtherServices_MRSN.aspx#HowMRSNWorks. The Antibacterial Resistance Leadership Group (ARLG) maintains a Virtual Biorepository at https://arlg.org/laboratory-center-strain-access. The National Institute of Allergy and Infectious Diseases (NIAID) maintains the Biodefense and Emerging Infections Research Resources Repository (BEI Resources), accessible at https://www.beiresources.org/. A list of international culture collections can be found at www.wfcc.info.
FUTURE PLANS FOR THE AR ISOLATE BANK
To fulfill the original objective of the AR Isolate Bank and to contribute further toward the goals set forth in the CARB initiatives, the AR Isolate Bank will continue to expand by adding more panels each year. We aim to further increase the phenotypic and genotypic diversity of bacterial isolates in the AR Isolate Bank. Isolates with novel and emerging resistance mechanisms will be added as they are detected. We will continue to develop and maintain international partnerships to obtain isolates with resistance mechanisms that have not yet been detected in the United States.
In addition to adding more isolates and panels, we are enhancing the AR Isolate Bank website to facilitate user navigation and enhance the ordering and shipping processes. The CDC and FDA will continue to perform WGS on all isolates in the AR Isolate Bank. WGS data will facilitate a more complete analysis and understanding of the mechanisms of resistance, including the correlation between genotypes and phenotypes. Such improvements to understanding genetic resistance mechanisms may contribute to better diagnostic capabilities. Genomic sequences can also support the development and evaluation of new antimicrobial agents.
The AR Isolate Bank is a publicly available resource that serves as a centralized source of challenge sets for device evaluation and validation. It is a useful tool that helps to fulfill the missions of both the CDC and FDA to enhance, promote, and protect public health. The AR Isolate Bank plays a facilitating role in many aspects of validation, verification, and regulatory review of new assays. Rapid and accurate detection of antimicrobial resistance allows for timely treatment with the appropriate antimicrobial agents and prompt implementation of recommended infection control measures.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Bell M. 2014. Antibiotic misuse: a global crisis. JAMA Intern Med 174:1920–1921. doi: 10.1001/jamainternmed.2014.3289. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention, Office of Infectious Diseases. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Office of Infectious Diseases, Atlanta, GA. http://www.cdc.gov/drugresistance/threat-report-2013 Accessed 19 May 2017.
- 3.Boucher HW, Bakken JS, Murray BE. 2016. The United Nations and the urgent need for coordinated global action in the fight against antimicrobial resistance. Ann Intern Med 165:812–813. doi: 10.7326/M16-2079. [DOI] [PubMed] [Google Scholar]
- 4.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 5.Jones RN. 1996. The emergent needs for basic research, education, and surveillance of antimicrobial resistance: problems facing the report from the American Society for Microbiology Task Force on Antibiotic Resistance. Diagn Microbiol Infect Dis 25:153–161. doi: 10.1016/S0732-8893(96)00099-5. [DOI] [PubMed] [Google Scholar]
- 6.O'Neil J. 2016. Tackling drug resistant infections globally: final report and recommendations. Review on Antimicrobial Resistance, London, England. https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf Accessed 31 May 2017.
- 7.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance 2014. World Health Organization, Geneva, Switzerland. http://www.who.int/drugresistance/documents/surveillancereport/en/ Accessed May 19, 2017.
- 8.The White House. 2014. Executive Order—combatting antibiotic-resistant bacteria. The White House, Washington, DC. https://obamawhitehouse.archives.gov/the-press-office/2014/09/18/executive-order-combating-antibiotic-resistant-bacteria. Accessed 19 May 2017.
- 9.The White House. 2015. National action plan to combat antibiotic-resistant bacteria. The White House, Washington, DC: https://obamawhitehouse.archives.gov/the-press-office/2015/03/27/fact-sheet-obama-administration-releases-national-action-plan-combat-ant. Accessed 19 May 2017. [Google Scholar]
- 10.FDA. 2009. Guidance for industry and FDA. Class II special controls guidance document: antimicrobial susceptibility test (AST) systems. FDA, Silver Spring, MD. http://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuGuida/UCM388961.pdf Accessed 19 May 2017.
- 11.FDA. 2014. Highly multiplexed microbiological/medical countermeasure in vitro nucleic acid based diagnostic devices: guidance for industry and Food and Drug Administration staff. FDA, Silver Spring, MD: https://www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidancedocumengu/ucm327294.pdf Accessed 19 May 2017. [Google Scholar]
- 12.FDA. 2016. Microbiology data for systemic antibacterial drugs. Development, analysis, and presentation: guidance for industry. Available at http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM182288.pdf Accessed 19 May 2017.
- 13.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomics sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.CLSI. 2017. Performance standards for antimicrobial susceptibility testing; 27th informational supplement. CLSI document M100-S27. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 15.CLSI. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard; 10th edition. CLSI document M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]