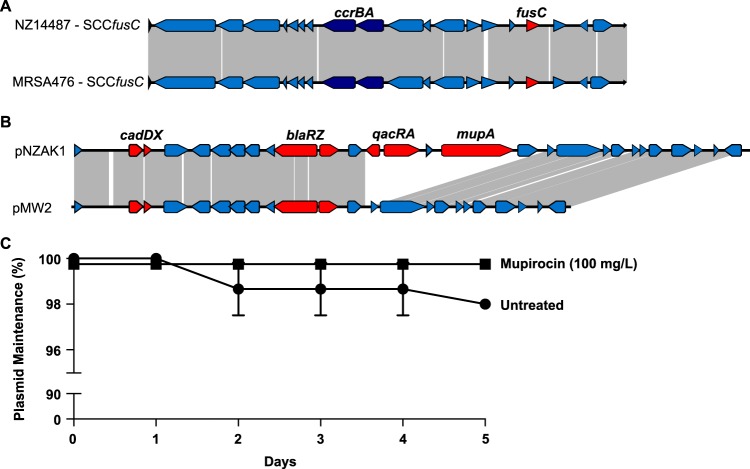
FIG 1.
(A) Schematic diagram illustrating the genetic organization of the SCCfusC region of NZ14487 in comparison to the fusC-harboring SCC476 element of MRSA476 (GenBank accession no. NC_002953). Regions of similarity are joined by gray lines. The fusC and ccrBA genes are highlighted. (B) Schematic diagram depicting the linear genomic comparisons of plasmid pNZAK1 from S. aureus strain NZ14487 and plasmid pMW2 from S. aureus strain MW2 (GenBank accession no. AP004832). Regions of DNA similarity are joined by gray lines. The location of predicted antimicrobial resistance genes (cadDX, blaRZ, qacRA, and mupA) are shown with red arrows. (C) Segregational stability of plasmid pNZAK1 in Staphylococcus aureus NZ14487. Cultures of S. aureus NZ14487 were serially passaged in either nonselective BHI broth or in broth supplemented with 100 mg/liter mupirocin. Plasmid loss was determined at the days indicated. The mean of triplicate independent cultures grown under each test condition are shown, with error bars representing the standard error of the mean (SEM).