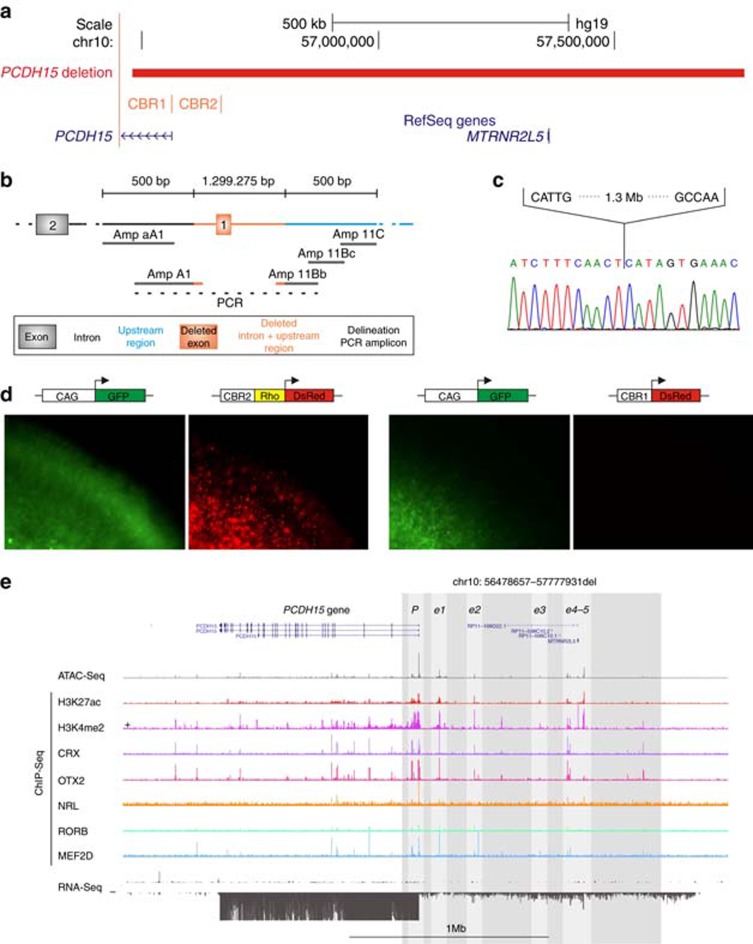
Figure 1.
Identification and characterization of a noncoding PCDH15 deletion. (a) Location of the deletion. The homozygous 1.3-Mb deletion identified in P9 is depicted in red, removing the first noncoding exon of the PCDH15 gene, the MTRNR2L5 gene with unknown function and two PCDH15-associated CRX-bound regions (CBR). CBR1 is situated in the promoter region, while CBR2 is located approximately 100 kb upstream. (b) Delineation of the deletion. Further refinement of the deletion breakpoint regions by conventional PCR. Black, orange and blue shaded boxes and connecting lines indicate nondeleted exon and introns, deleted exon, intron and upstream region, and nondeleted upstream region, respectively. Short grey horizontal lines correspond to designed PCR amplicons, used to delineate the deletion. The black dotted line indicates the junction product. (c) Sanger sequencing of the junction product. Delineation of the deletion at the nucleotide level, chr10: g.56478660_57777934del. (d) Electroporation assays. In order to assess the cis-regulatory effect of CBR1 and CBR2, electroporation reporter assays were performed in mouse retinal explants. The first construct consisted of CBR2 cloned in a dsRed expressing vector, upstream of a Rho-basal promoter, as CBR2 is located in a more distant regulatory region. For the second construct, CBR1 was cloned in a dsRed expressing vector without basal promoter, as it is located in the promoter region. Cis-regulatory activity could be demonstrated for the CBR2 construct, while CBR1 seems to fail in driving dsRed expression on its own. (e) Cis-regulatory landscape of PCDH15. Epigenomic marks and transcription factor binding were assessed in human adult retina, shown here for the PCDH15 locus: ATAC-seq; ChIP-seq for H3K27ac, H3K4me2, CRX, OTX2, NRL, RORB and MEF2D; and RNA-seq. The region of the deletion is marked with a shaded rectangle and the putative active promoter and enhancer regions included in the deleted region are in lighter shading.