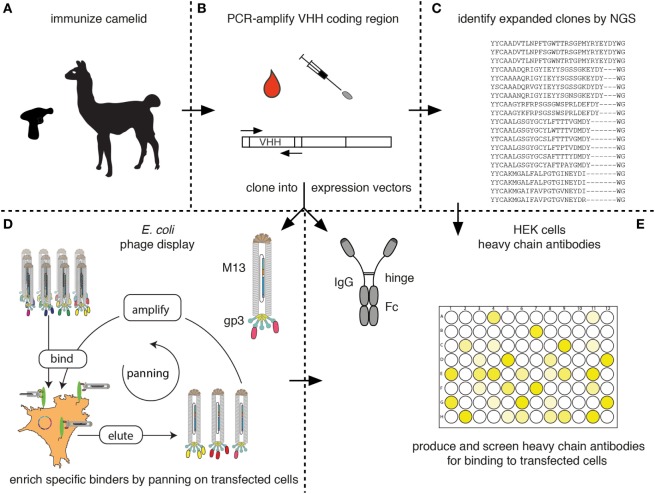
Figure 7.
Schematic diagram for the identification of membrane protein-specific nanobodies (Nbs). This figure schematically illustrates the discovery path for membrane-protein-specific Nbs following cDNA immunization. (A) Camelids are immunized as described in Figure 5. (B) RNA is prepared from lymphocytes obtained from blood or a lymph node biopsy and transcribed into cDNA. The Nb (VHH) coding region is PCR amplified. (C) An aliquot of the PCR product is subjected to next-generation sequencing. The results permit identification of expanded clones and reveal the extent of somatic hypermutation in such clones. (D,E) A second aliquot of the PCR product is cloned into a bacterial (D) and/or mammalian (E) expression vector. (D) During cloning into the bacterial vector, the Nb is genetically fused to the gp3 capsid protein of the M13 phage. Antigen-specific clones can then be enriched from the phage display library by panning on cells transfected with the membrane protein of interest. Bound phage are eluted and panning can be repeated, e.g., on another cell type transfected with the protein of interest. Panning can be performed in the presence of available antibodies or Nbs in order to select for Nbs binding to a distinct epitope. (E) During cloning into the mammalian vector, the Nb is genetically fused to the hinge and Fc domains of a conventional rabbit, mouse, or human IgG. Individual clones are transfected into HEK cells. Heavy-chain antibodies secreted into the HEK cell supernatant are then screened for binding to cells transfected with the membrane protein of interest by ELISA or immunofluorescence microscopy using appropriate enzyme- or fluorochrome-conjugated secondary antibodies.