Fig. 4.
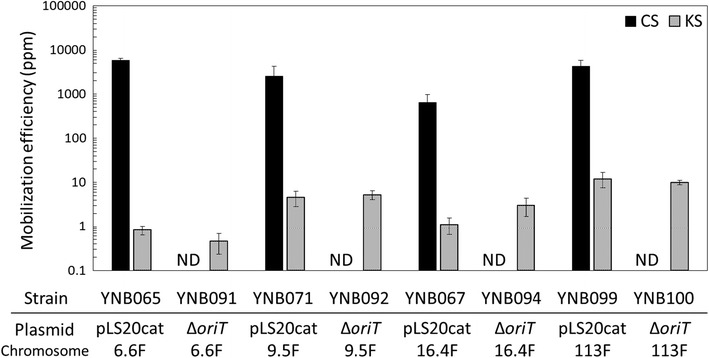
Mobilization efficiencies of the kanamycin resistance gene at the aprE locus and the helper plasmids, pLS20cat and pLS20catΔoriT. Liquid cultures of the recipient strain YNB001 (comK::spc) and one of the donor strains: YNB065 (YNB060 with pLS20cat), YNB091 (YNB060 with pLS20catΔoriT), YNB071 (YNB069 with pLS20cat), YNB092 (YNB069 with pLS20catΔoriT), YNB067 (YNB062 with pLS20cat), YNB094 (YNB062 with pLS20catΔoriT), YNB099 (YNB097 with pLS20cat), and YNB100 (YNB097 with pLS20catΔoriT), were mixed for conjugative transfer of long DNA segments (6.6F, 9.5F, 16.4F, and 113F for 6.6, 9.5, 16.4, and 113 kb between oriTLS20 and the kanamycin marker, respectively), and spread on LB plates containing both chloramphenicol and spectinomycin (CS), both kanamycin and spectinomycin (KS), and spectinomycin alone. Colonies were counted as CFUs to calculate mobilization efficiencies [CFU of transconjugants (colonies on the CS and KS plates)/CFU of total recipients (colonies on the spectinomycin plate) × 106 (ppm)]. Values are means with standard deviations from three independent experiments. ND not detected (< 0.01 ppm)
