Extended Data Figure 6. Gsdmd expression in different tissues and generation of Gsdmd deficient mice.
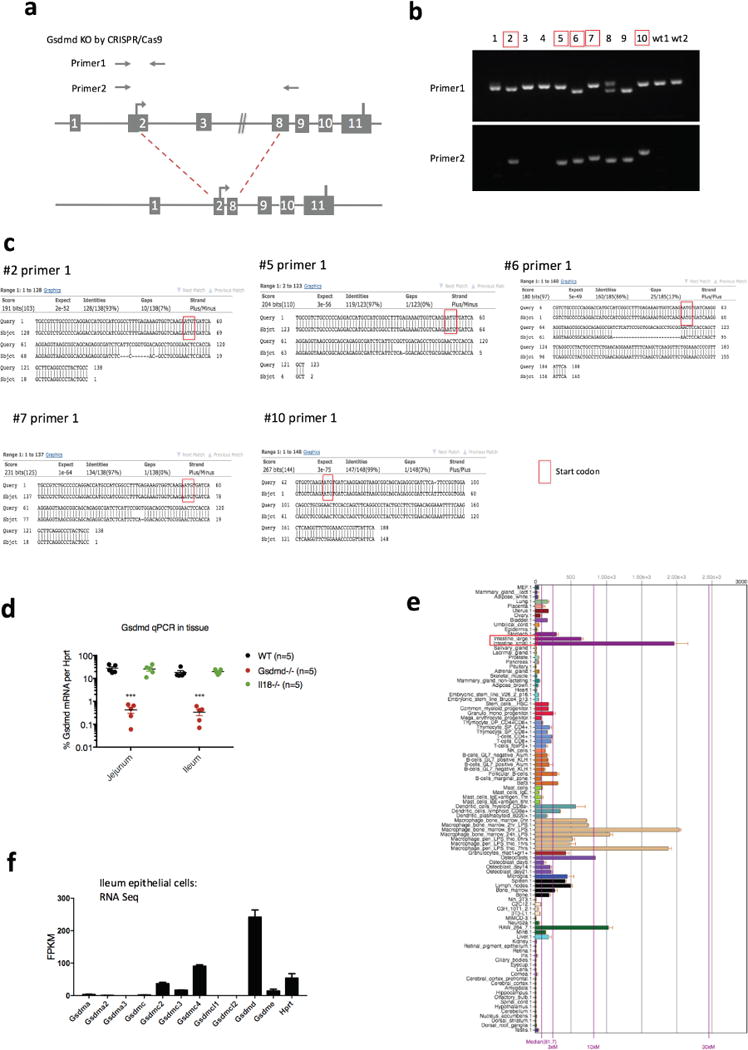
a, Illustration of the CRISPR–Cas9 strategy to generate Nlrp9b-deficient mice and primer design used in b. b, Genotyping of the Gsdmd mutant pups. The mice chosen for experiments shown in Fig. 3c, d are shown in red, with a deletion in one allele sequenced by primer 2, as well as mutations that result in frame shift in another allele sequenced by primer 1. c, Sequencing of redlabelled mice with primer 1. d, qPCR validation of Gsdmd expression in the intestine tissues from these five red-labelled Gsdmd mutant mice, comparing with wild-type mice and Il18−/− mice (mean ± s.e.m., Student’s t-test, ***P < 0.001). e, Gsdmd expression in different mouse tissues as per BioGPS. f, RNA sequencing data reveals the expression of gasdermin family members and housekeeping gene Hprt in the ileal epithelial cells of wild-type mice. b–d, Representative of two experiments and the data was combined in Fig. 3c and d.
