Fig. 1. Assembly of the Chl biosynthesis pathway in Escherichia coli.
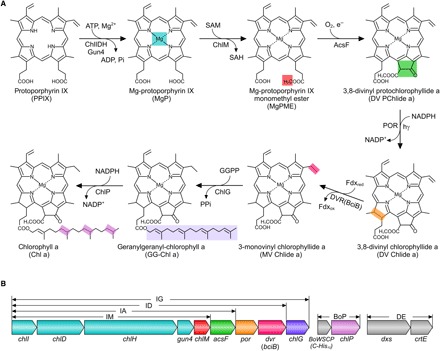
(A) Overall reactions from PPIX to Chl a catalyzed by the enzymes introduced to E. coli. The insertion of Fe2+ into PPIX (not shown) creates a biosynthetic branchpoint (not shown) that yields heme. Colored shading denotes the chemical change(s) at each step. ATP, adenosine triphosphate; ADP, adenosine diphosphate; SAM, S-adenosine-l-methionine; SAH, S-adenosyl-l-homocysteine; NADP+, nicotinamide adenine dinucleotide phosphate; NADPH, reduced form of NADP+; Pi, inorganic phosphate; PPi, inorganic pyrophosphate. (B) Arrangement and relative size of each gene in the constructed plasmids. The chlI, chlD, chlH, gun4, chlM, acsF, dvr (bciB), and chlG genes were consecutively cloned into a modified pET3a vector with a single T7 promoter upstream of chlI and a ribosome-binding site upstream of each gene using the link-and-lock method (see fig. S2 for details) (40). Colors for genes correspond to those used in (A). Plasmid constructs and gene contents are IM (chlI–chlM), IA (chlI–acsF), ID (chlI–dvr), IG (chlI–chlG), BoP (BoWSCP and chlP), and DE (dxs and crtE). BoP is a pACYCDuet1-based plasmid containing a sequence encoding the BoWSCP protein with a C-terminal His10 tag (10) and the Synechocystis chlP gene. DE is a pCOLADuet1-based plasmid containing the E. coli dxs gene and the Rvi. gelatinosus crtE gene.
