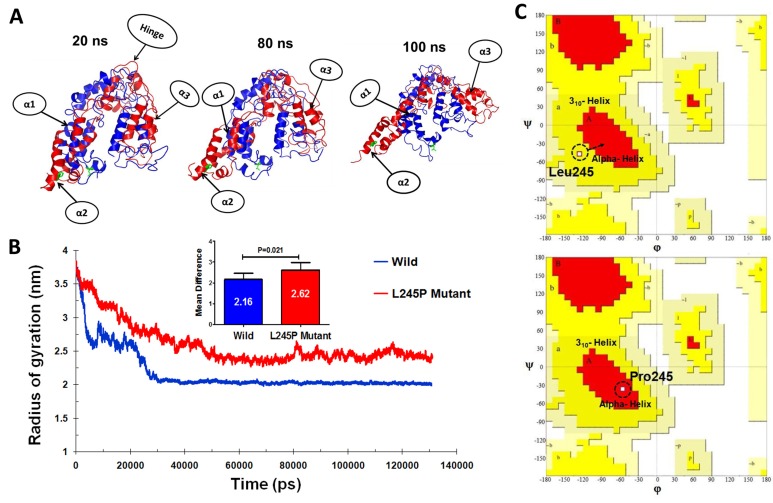
Figure 4. Tertiary structural stability of the (c.734T > C) p.L245P mutant PROM1.
A. Structural superposition of the wild- (red) and the mutant-type (blue) domains extracted from the trajectories of 20, 80, and 100 ns. The mutation of L245P and the residue Leu245 of wild type are labeled in green color. The helices and hinge indicate with arrows. α1: residues number 70-77, α2: bHLH-Zip domain, and α3: residues number: 183-191. B. Plot of average RG values for each of the systems. RG for L245P mutant and wild type are colored as red and blue, respectively. The chart also shows differential RG values between residues of WT and mutant forms. There is a strong prospect that the L245P mutation influences the structure of the active site of PROM1, possibly by creating a further fold and destabilizing the mentioned domain. C. Comparative diagram depicting Ramchandran plot analysis of PROM1 protein variants in the wild type (upper panel) and the L245P mutant (lower panel) during 130 ns molecular dynamic simulations. Ramachandran plots show the phi (φ)-psi (ψ) torsion angles for the related residue number 245 of PROM1 in this structure. Leu and Pro residues are shown as square (□) and are not restricted to the regions of plots. In the upper panel, before mutation, Leu245 located in the 310 helix region, whereas mutated residue (L245P) transferred to the alpha helix parts of plot in the lower panel during the simulation time.