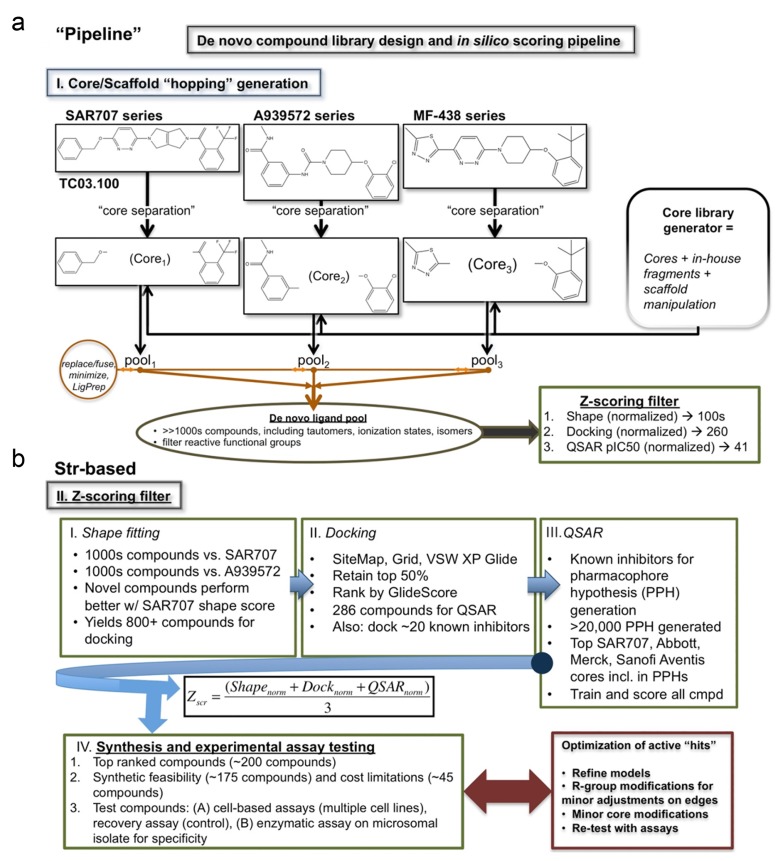
Figure 1. De novo compound library design and in silico scoring pipeline.
a. Core, or scaffold, hopping generation for three known commercial SCD1 inhibitors (SAR707, A939572, and MF-438) is shown. The central scaffold is separated from the compound (“core separation”) leaving the binding features from the edge of each compound. The core library generator then inserts new cores fuses the edges, minimizes the structure energy, and prepares the ligands (LigPrep). The de novo ligands are then pooled and screened for reactive functional groups. The final set of compounds is then fed into our reductive Z-scoring filter. b. Structure-based reductive filter for SCD1 specific compounds. The Z-scoring filter operates in three iterative steps: Shape filter, Docking filter, and QSAR filter. The shape filter generates 100s of conformers for each of the thousands of compounds generated to best fit either SAR707 or A939572. Best fit of compounds with SAR707 had most surviving compounds (>800), thus selected for next filter, docking. Docking filter was applied to >800 compounds from the shape filter. Glide-XP docking retaining top 50% and addition of known inhibitors yields a pool of 286 compounds for QSAR filtering. A QSAR model was made from over 20,000 pharmacophore hypotheses based on a set of 32 known compounds ranging from low nanomolar activity to high milimolar (no activity). The QSAR model trained on this dataset generating a final pool of compounds with good predicted IC50. The algorithm for ranking these final 242 compounds is shown. A subset of 38 compounds from the Z-score filtering was selected for synthesis and testing. Compounds were then tested with cell-based assay and enzymatic assay for activity and specificity. Optimization for these compounds may proceed as needed.