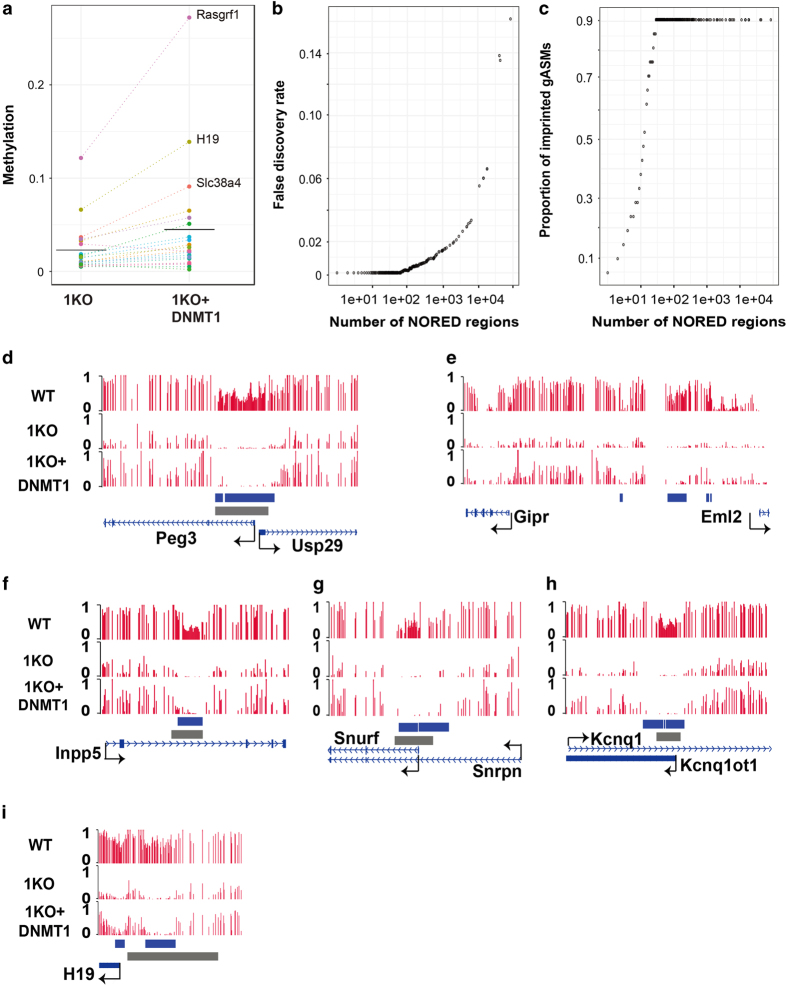
Figure 2.
Loss of methylation was not rescued at gASMs and other specific loci. (a) Profiling of gASM methylation level in 1KO and DNMT1-rescued 1KO (r1KO) ESCs. For most gASMs, low methylation in 1KO is not substantially increased by exogenous expression of DNMT1 cDNA in r1KO ESCs. Genome-wide, average CpG methylation on a zero to one scale was 0.369 for r1KO cells. See Supplementary Table S1 for locus-specific details. (b) False discovery rate (FDR, y axis) of NORED as a function of rank (x axis) for NORED regions. (c) Proportion of well-established gASMs (y axis) identified by NORED as a function of rank (x axis) for NORED regions. Nineteen gASMs rank among top 29 NORED regions. (d) Deficiency of methylated CpG sites (red bars) in 1KO and DNMT1-rescued 1KO cells can be used to identify and demarcate the known Peg3 DMR. (e) NORED identifies Gipr/Eml2 locus (imprinted status unknown). (f) NORED identifies, gASM in Inpp5f locus. (g) NORED improves the demarcation of gASMs for Prader-Willi and Angelman syndromes (Snrpn/Snurf). (h) NORED demarcates region larger than known gASM at Kcnq1/Kcnqot1 locus. (i) NORED successfully identifies gASM and somatic ASM at near H19. (d–i) Blue bars indicate NORED regions. Gray bars indicate location of previously established gASMs.