Summary
Double‐stranded RNA (dsRNA) molecules targeting two genes have been identified that suppress economically important parasitic nematode species of banana. Proteasomal alpha subunit 4 (pas‐4) and Actin‐4 (act‐4) were identified from a survey of sequence databases and cloned sequences for genes conserved across four pests of banana, Radopholus similis, Pratylenchus coffeae, Meloidogyne incognita and Helicotylenchus multicinctus. These four species were targeted with dsRNAs containing exact 21 nucleotide matches to the conserved regions. Potential off‐target effects were limited by comparison with Caenorhabditis, Drosophila, rat, rice and Arabidopsis genomes. In vitro act‐4 dsRNA treatment of R. similis suppressed target gene expression by 2.3‐fold, nematode locomotion by 66 ± 4% and nematode multiplication on carrot discs by 49 ± 5%. The best transgenic carrot hairy root lines expressing act‐4 or pas‐4 dsRNA reduced transcript message abundance of target genes in R. similis by 7.9‐fold and fourfold and nematode multiplication by 94 ± 2% and 69 ± 3%, respectively. The same act‐4 and pas‐4 lines reduced P. coffeae target transcripts by 1.7‐ and twofold and multiplication by 50 ± 6% and 73 ± 8%. Multiplication of M. incognita on the pas‐4 lines was reduced by 97 ± 1% and 99 ± 1% while target transcript abundance was suppressed 4.9‐ and 5.6‐fold. There was no detectable RNAi effect on nontarget nematodes exposed to dsRNAs targeting parasitic nematodes. This work defines a framework for development of a range of nonprotein defences to provide broad resistance to pests and pathogens of crops.
Keywords: RNA interference, nematode resistance, double‐stranded RNA, transgenic hairy roots, banana, crop
Introduction
Cooking bananas are an important staple crop for 400 million people in low‐ and middle‐income countries throughout the tropics; in Africa, 70 million people rely on them for more than 25% of their carbohydrates (FAOSTAT, 2014; Ortiz and Vuylsteke, 1996). Annual production of 139 million tons is predominantly by small‐scale farmers to feed their families and for sale at local markets (FAOSTAT 2014). Consequently, the crop is not subject to the same price shocks as other globally traded staples such as rice and wheat (West et al., 2014). In tropical regions, where the crop is grown, cooking banana is also capable of feeding more people cheaply per unit area than other staple crops (Frisson and Sharrock, 1999), as such it has an important role to play in ensuring food security in regions of the world that suffer from chronic malnutrition.
Banana is a sterile, genetically homogeneous crop that is severely hampered by several pests and pathogens. In particular, banana plantations can suffer over 50% production losses per crop cycle as a result of plant‐parasitic nematode infections (Norgrove and Hauser, 2014; Roderick et al., 2012; Ssango et al., 2004). Control of the nematode species complexes of Radopholus similis, Pratylenchus goodeyi, P. coffeae, Helicotylenchus multicinctus and Meloidogyne spp. is required across many banana‐growing regions (Bridge et al., 1995; Gowen et al., 2005; Kashaija et al., 1994). Growers are frequently not aware of the importance of nematodes or the species present in their plantations (Hauser and Amougou, 2010). A lack of effective resistance genes and sterility both limit progress by conventional breeding, but sterility also prevents gene flow, strongly favouring a transgenic approach for nematode resistance (Lorenzen et al., 2010).
RNA interference (RNAi) has great potential for the development of transgenic crops with resistance to a range of pests and pathogens, as reviewed by Kaur et al. (2016). The RNAi effect was first described for Caenorhabditis elegans (Fire et al., 1998) and subsequently for plant‐parasitic nematodes (Urwin et al., 2002). Plants expressing dsRNA targeting nematode genes have been shown to suppress Heterodera schachtii (Patel et al., 2008, 2010; Sindhu et al., 2009), H. glycines (Klink et al., 2009; Li et al., 2010), Meloidogyne sp. (Huang et al., 2006; Ibrahim et al., 2011; Yadav et al., 2006) and R. similis (Li et al., 2015a,b). Additionally, in vitro treatment of P. coffeae, P. thornei and P. zeae with dsRNA targeting troponin C (pat‐10) or a calponin (unc‐87) resulted in aberrant movement and reduced multiplication on carrot discs (Joseph et al., 2012; Tan et al., 2013).
Optimization of dsRNA is required to both ensure that the intended target genes are silenced and to decrease the likelihood of off‐target effects. The exact level of conservation of sequence required to initiate RNAi has yet to be clearly defined. Exact matches between siRNAs generated from a dsRNA and target transcript over 19 nucleotides (nt) have been reported as sufficient for silencing (Naito et al., 2005), although off‐target effects in mammals can be triggered by a 6‐nt seed at the 5′ end of the siRNA (Kamola et al., 2015). A study that progressively reduced the homology between dsRNA and target sequence suggested that there is a gradual reduction in efficacy rather than a sharp loss of effect (Parrish et al., 2000). Another potential aspect of dsRNA molecules that has been shown to impact efficacy is molecule length. Molecules may have to be at least 50 bp long in insects, with no effect when a 25‐bp molecule is used (Yang and Han, 2013). The position of the region targeted by a dsRNA may also affect the efficacy of RNAi. Molecules designed to target 3′ regions of a transcript have proven more potent than targeting 5′ regions in nematodes (Lilley et al., 2012), although this is not necessarily true in insects (Yang and Han, 2013). To control across species, sequence conservation of the target region is also an important consideration. Experimental testing of any in silico prediction is necessary because the criteria for RNAi efficacy are not fully defined.
The RNAi approach potentially reduces regulatory hurdles as it involves no expression of novel protein or peptide thereby enhancing its inherent biosafety (Parrott et al., 2010), although for large‐scale production and commercialization, a range of biosafety assessments are still required (Auer and Frederick, 2009; Heinemann et al., 2013; Roberts et al., 2015). The level of food and feed safety of RNAi defences is likely to be high due to extensive degradation, biological barriers to uptake and low efficacy of exogenous nucleic acids ingested by mammals (Petrick et al., 2013). Environmental risks to nontarget organisms such as nematodes and insects that feed on the transgenic plant can be managed. This requires appropriate genetic information to avoid of dsRNA sequences likely to induce off‐target effects (Rual et al., 2007; Yamada and Morishita, 2005). Alternative approaches are required when sequence of putatively at‐risk species is not available from databases. One strategy involves assessing cross‐hybridization of dsRNA molecules to representative model species with well‐characterized transcriptomes (Naito et al., 2005). A second is exposure of sentinel, environmental indicator species that are highly RNAi susceptible to the dsRNA (Custodia et al., 2001; Winston et al., 2007).
There is a great need for a rational defence against a proscribed range of pathogens without adverse effects on nontarget organisms. In this study, the control of R. similis and P. coffeae were prioritized as the most damaging pests of banana (Gowen et al., 2005) and Meloidogyne introduced to test broader resistance conferred by the dsRNA molecules of interest that had been identified in the initial phase. The dsRNA molecules identified in this study offer a first step in realizing the potential for a broadly based resistance to economically important nematode pests in an important staple crop. The approach is readily extendable to other crops with a range of nematode pests such as rice. Additionally, transformation of crop plants, and particularly banana, involves a considerable investment in both time and resources. We address this constraint by describing a framework to identify and validate biosafe dsRNA molecules that have the potential to enhance the rate of development of future RNAi defences for many crop pests and pathogens.
Results
Target identification
Bioinformatic comparison of C. elegans genes with severe RNAi phenotypes and conservation to the economically important banana nematodes R. similis, P. coffeae, H. multicinctus and Meloidogyne incognita identified nine genes that shared >80% sequence identity between C. elegans and at least two of the banana nematodes. Sequences for C. elegans, R. similis and M. incognita were obtained from publically available databases. Amplification from cDNA was used to obtain sequences for P. coffeae and H. multicinctus using primers designed to conserved regions identified from database sequences (Table 1). Sequences with sufficient conservation for dsRNA design across all four target nematodes were identified for Actin‐4 (act‐4) and proteasomal alpha subunit 4 (pas‐4) genes (Table 2). Double‐stranded RNA (dsRNA) molecules were designed to target regions of these two genes that had one or more exact 19 nucleotide sequence matches. These regions corresponded to a 376‐bp section within the substrate‐binding domain region of ACT‐4 and a 292‐bp section of the PAS‐4 N‐terminal nucleophile (Ntn)‐hydrolase active site. The sequences used were derived from cDNA of P. coffeae for pas‐4 and H. multicinctus for act‐4 (Figure S1). A search for potential off‐target effects of the two dsRNA sequences was completed using a database (http://dsCheck.RNAi.jp/; Naito et al., 2005). It identified 19mers matching five C. elegans genes, eight Drosophila genes, five rat genes and one rice gene. All C. elegans and rat matches were eliminated by truncating the act‐4 sequence to 112 bp (act‐4_t). A 19mer match to one rice gene and matches to two Drosophila genes were still present in the truncated act‐4 sequence. There were no nontarget organism 19mer matches to the pas‐4 dsRNA molecule.
Table 1.
Potential target genes identified from database sequences
| Gene | Wormbase ID | C. elegans expression | C. elegans RNAi phenotype | Rs | Mi | Pc | Hm |
|---|---|---|---|---|---|---|---|
| ACT‐4 |
WBGene 00000066 |
Body wall and vulval muscles | Embryonic lethal | RSC00784 | MIC00350 | ✓ | ✓ |
| CCO‐1 |
WBGene 00000371 |
Mitochondrial | Lethal | RSC00550 | – | ✓ | – |
| HSP‐60 |
WBGene 00002025 |
Mitochondrial | Embryonic lethal | RSC02271 | MIC03015 | – | – |
| PAS‐3 |
WBGene 00003924 |
All major muscles | Embryonic lethal | RSC00234 | MIC05526 | ✓ | – |
| PAS‐4 |
WBGene 00003925 |
Pharynx and body wall muscle | Lethal | ✓ | MIC06652 | ✓ | ✓ |
| RPS‐23 |
WBGene 00004492 |
Muscles and nervous system | Embryonic lethal | RSC00184 | MIC03023 | – | – |
| UBQ‐1 |
WBGene 00006727 |
Muscles, hypodermis and nervous system | Lethal | RSC02466 | MIC06345 | – | ✓ |
| UBQ‐2 |
WBGene 00006728 |
Muscles and nervous system | Embryonic lethal | RSC00037 | MIC06345 | ✓ | – |
| UNC‐87 |
WBGene 00006819 |
All major muscles | Lethal | RSC04225 | MIC03833 | – | – |
RNAi phenotypes are the most severe given on Wormbase for C. elegans. The Nembase 4 gene reference is given for R. similis and M. incognita sequences obtained following a BLAST search with the C. elegans sequence. Sequences amplified from cDNA either had >80% sequence identity and 21‐nt regions with (✓) or without (–) 100% identity to other banana nematode sequences. The plant‐parasitic nematodes are as follows: R. similis (Rs), M. incognita (Mi), Pratylenchus coffeae (Pc) and Helicotylenchus multicinctus (Hm).
Table 2.
Sequence identity of act‐4 and pas‐4 dsRNA molecules to target gene sequences of five nematodes with the species to which they were cloned indicated (ex‐)
| act‐4 (ex H. multicinctus) | pas‐4 (ex P. coffeae) | ||
|---|---|---|---|
| Nematode | 376 bp | 112 bp | 307 bp |
| Radopholus similis | 90% | 88% | 100% |
| Helicotylenchus multicinctus | 100% | 100% | 100% |
| Pratylenchus coffeae | 90% | 79% | 100% |
| Meloidogyne incognita | 81% | 87% | 96% |
| Caenorhabditis elegans | 86–83% (5) | 81–85% (5) | 68% |
Value ranges of sequence similarities are for the number of orthologues, as indicated in parentheses. The dsRNA sequence similarities to targeted genes are subdivided into three categories: (i) at least one 21 perfect nucleotide sequence match (no shading), (ii) one or more 40 nucleotide sequences that share 38–39 nucleotides (lighter grey shading) and (iii) other, lower levels of similarity (darker grey shading).
In vitro dsRNA treatment
Quantitative RT‐PCR established that dosing with dsRNA for 24 h resulted in a significant 1.5‐fold reduction of pas‐4 transcript abundance (P < 0.05, one‐way ANOVA with Tukey Post hoc) for R. similis and 2.3‐fold (P < 0.001) for P. coffeae relative to the gfp dsRNA control treatment. Both the act‐4 long dsRNA and truncated dsRNA molecules significantly suppressed act‐4 transcript in R. similis by 2.3‐ and 1.9‐fold, respectively (P < 0.001), but neither had a significant effect on P. coffeae transcript levels (Figure 1). The act‐4 targeting molecules decreased the rate of locomotion of R. similis by 79 ± 9% for the 376‐bp molecule and 66 ± 4% for the 112‐bp molecule (P < 0.001) relative to control nematodes (Figures 2a and Video S1). The longer act‐4 molecule also reduced the locomotion of P. coffeae by 64 ± 7% (P < 0.001), but the truncated version did not (Figures 2b and Video S1). Targeting pas‐4 had no significant effect on the rate of locomotion of either nematode (Video S1).
Figure 1.
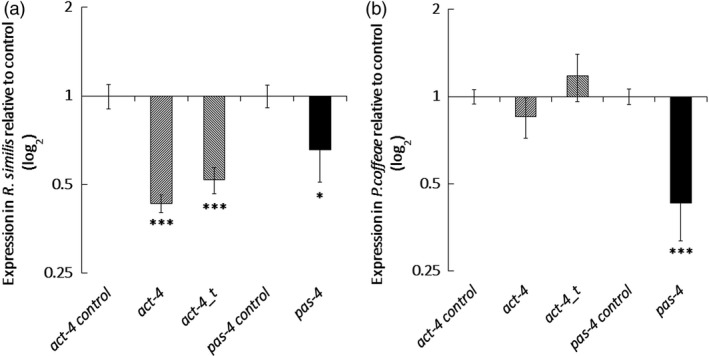
Quantitative RT‐PCR Measurement of Transcript Knock‐down. Reduction is for act‐4 and pas‐4 expression following soaking in 100 μg/mL dsRNA for 24 h for (a) Radopholus similis and (b) Pratylenchus coffeae. Values are means ± SEM for four technical replicates of three biological replicates with significant loss of expression relative to nontargeting gfp dsRNA treatment indicated. Analysis is based on univariate ANOVA of log values with Tukey post hoc comparison to gfp, ***P < 0.001; *P < 0.05.
Figure 2.
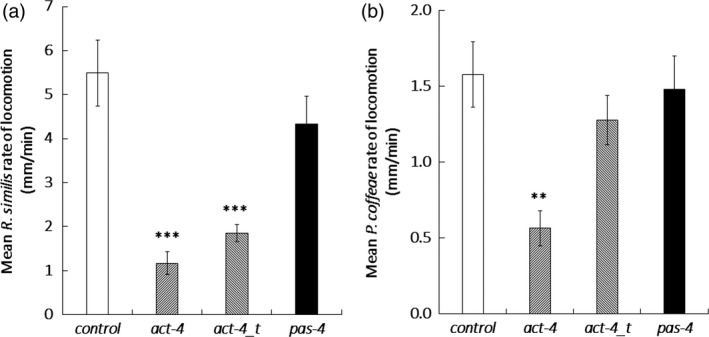
Effect of dsRNA treatment on nematode locomotion. Speed of locomotion (mm/min) was recorded for (a) Radopholus similis and (b) Pratylenchus coffeae on an agar surface after soaking in 100 μg/mL dsRNA targeting act‐4 or pas‐4. Significance is given from post hoc comparison of each RNAi effect relative to the gfp dsRNA control. Analysis is based on univariate ANOVA of log values (n > 15) with Tukey post hoc comparison to gfp, ***P < 0.001; **P < 0.01.
Multiplication of R. similis and P. coffeae on carrot discs
Long‐term effects of the three dsRNA molecules were studied by weekly dosing during culture on carrot discs over 8 weeks. The long and truncated act‐4 dsRNA molecules suppressed R. similis population increase by 72 ± 11% (P < 0.001) and 49 ± 5% (P < 0.01), respectively, and the pas‐4 targeting dsRNA by 89 ± 2% (P < 0.001) relative to controls (Figure 3a). R. similis nematodes recovered from the carrot discs at 8 weeks also had significant reduction in targeted transcript (Figure 3c) No significant reduction occurred in multiplication or transcript levels of P. coffeae individuals with any of the three dsRNA treatments (Figure 3b,c).
Figure 3.
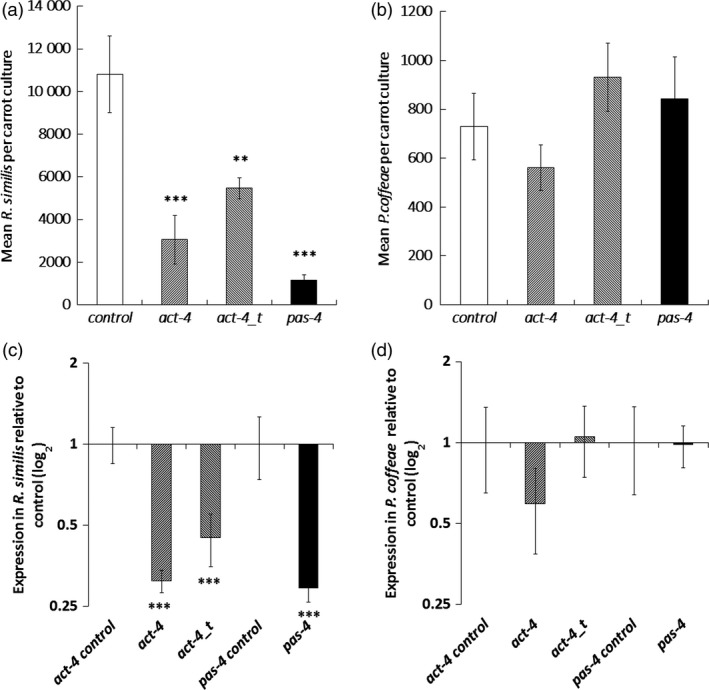
Effect of dsRNA treatment of nematode cultured on carrot discs. The effect of dosing nematodes weekly with dsRNA molecules on (a,b) mean population density and (c,d) target transcript level was assessed after 8‐week culture on carrot discs for (a,c) Radopholus similis and (b,d) Pratylenchus coffeae. Dosing was with dsRNA molecules in M9 buffer targeting sequences of act‐4 (act‐4 and act‐4_t) or pas‐4. Quantitative RT‐PCR values are means ± SEM for four technical replicates of three biological replicates with significant loss of expression relative to nontargeting gfp dsRNA treatment indicated. Analysis is based on univariate ANOVA of log values (n = 10) with Tukey post hoc comparison to gfp, ***P < 0.001; **P < 0.01.
Transgenic hairpin carrot hairy roots
The in planta suppression of R. similis, P. coffeae and M. incognita when targeting act‐4 or pas‐4 was determined using transgenic carrot hairy roots expressing hairpin constructs for dsRNA production. An initial challenge with R. similis was carried out with three replicates of 10 RT‐PCR‐positive independent transformation events per construct, including a dsRNA gfp construct as a null target control. This screen identified four lines, act‐4_t lines 5 and 6, pas‐4 lines 3 and 4, which were most effective at suppressing nematode multiplication. Nine act‐4_t lines and four pas‐4 lines provided >50% resistance. Additionally, two control gfp lines were chosen at random for subsequent challenges.
Nematode multiplication (Figure 4a) was significantly reduced (P < 0.001) for R. similis by 93 ± 1% and 94 ± 2% on act‐4_t lines 5 and 6 and 46 ± 6% and 69 ± 3% on pas‐4 lines 3 and 4. Transcript levels of the targeted genes (Figure 4b) in R. similis were also significantly suppressed (P < 0.001) 7.7‐fold and 7.9‐fold for act‐4_t lines 5 and 6 and 3.7‐fold and fourfold for pas‐4 lines 3 and 4. Multiplication of P. coffeae was also significantly reduced (P < 0.01) on all four lines, 42 ± 8% and 50 ± 6% on act‐4_t lines and 56 ± 8% and 73 ± 8% on pas‐4 lines. Nematode transcript abundance for act‐4_t line 5 was 1.8‐fold reduced (P < 0.01) and 2.3‐fold and twofold for pas‐4 lines 3 and 4 (P < 0.001). There was no significant reduction in the number of M. incognita J2s recovered from either act‐4_t line while a small but significant 1.3‐fold reduction in transcript abundance was detected for line 5 (P < 0.01). In contrast, the two pas‐4 lines reduced the multiplication of this nematode by 97 ± 1% and 99 ± 1% (P < 0.001) and transcript abundance by 4.9‐ and 5.6‐fold (P < 0.001). There was a significant difference (P < 0.05) between the relative efficacy of the two constructs against the different nematode species. R. similis was more disrupted by act‐4_t whereas pas‐4 was more effective against P. coffeae and M. incognita.
Figure 4.
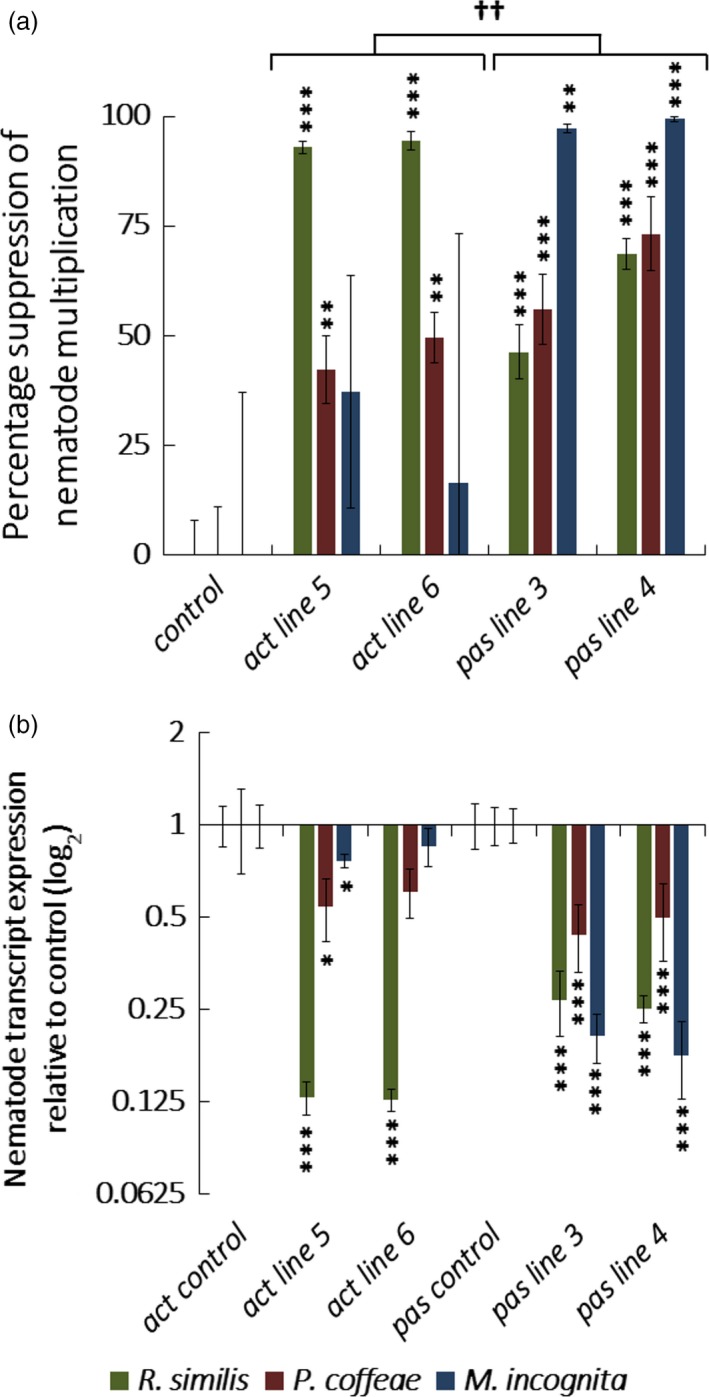
Transgenic Carrot Hairy Root Challenges. (a) Percentage suppression of multiplication obtained and (b) fold transcript knock‐down for nematodes recovered from hairy root lines expressing dsRNA targeting act‐4, pas‐4 or gfp after 10 weeks of culture when challenged with Radopholus similis, Pratylenchus coffeae or Meloidogyne incognita. Analysis is based on one‐way ANOVA of log values (n = 15) with Tukey post hoc comparison of individual lines to gfp control (***P < 0.001; **P < 0.01; *P < 0.05) and a priori ANOVA both act‐4_t lines to both pas‐4 lines (†† P < 0.01).
Effect on nontarget nematodes
A faunal analysis identified Caenorhabditis and Cephalobus species in soil samples from a transgenic banana field trial site at Kawanda, Uganda. Both species readily took up fluorescein isothiocyanate (FITC) in vitro (data not shown) and were selected as suitable sentinel species for an off‐target study. In vitro treatment with the act‐4 dsRNA molecules designed to target plant‐parasitic nematodes did not reduce locomotion of C. elegans in contrast to its counterpart directed at the endogenous act‐4 gene (Figure 5a). Both this dsRNA and one targeted at pas‐4 of C. elegans significantly reduced expression (P < 0.001; Figure 5b) in contrast to when this nematode was soaked in the banana nematode dsRNA variants. None of the dsRNA molecules targeted at either plant‐parasitic nematodes or C. elegans reduced the rate of locomotion of Cephalobus (Figure 5c).
Figure 5.
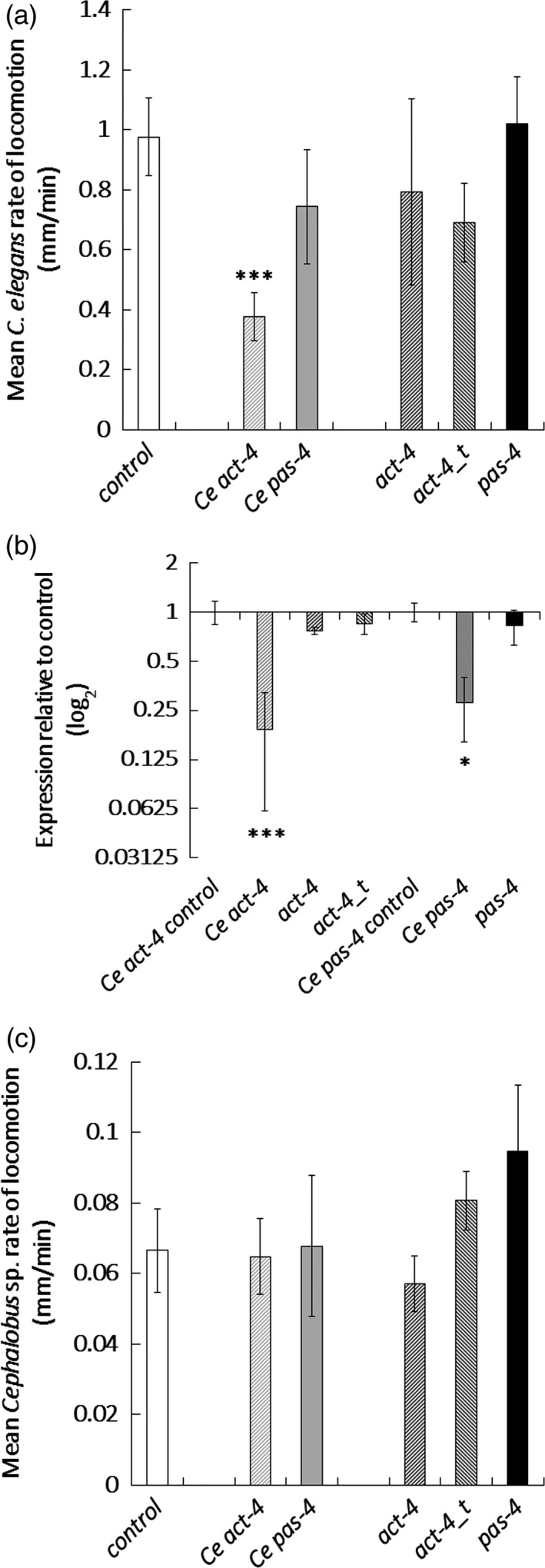
Effect of dsRNA treatment on nontarget nematodes. (a) Mean rate of locomotion and (b) transcript knock‐down was recorded for active Caenorhabditis elegans nematodes soaked for 24 h in 100 μg/mL dsRNA targeting sequences specific to C. elegans (Ce) or plant‐parasitic nematodes and (c) mean rate of locomotion obtained for Cephalobus sp. soaked for 24 h in 100 μg/mL dsRNA targeting sequences specific to C. elegans (Ce) or plant‐parasitic nematodes. Analysis is based on univariate ANOVA of log values (n > 15) with Tukey post hoc comparison to gfp, ***P < 0.001; *P < 0.05.
Discussion
This work identifies dsRNA sequences capable of cross‐genera control of economically important crop pests. Transcripts that have lethal phenotypes when knocked out in C. elegans and conservation across plant‐parasitic nematodes were targeted with dsRNA in vitro and in vivo. Control was achieved by targeting pas‐4, a proteasome alpha‐type seven subunit of the core 20S proteasome subcomplex involved in protein turnover (Papaevgeniou and Chondrogianni, 2014), and act‐4, an actin gene fundamental to muscle function. Both genes are predominantly expressed in C. elegans body wall muscles.
Targeting pas‐4 suppressed parasitism and transcript on transgenic hairy roots for R. similis, P. coffeae and M. incognita. The effectiveness of the pas‐4 dsRNA molecule when expressed in planta against all three banana nematodes may be due to its high conservation between all plant‐parasitic nematodes. The lack of a significant effect in vitro on P. coffeae may reflect that weekly dosing with dsRNA on carrot discs is insufficient to maintain the RNAi effect achieved relative to continual exposure in vivo. By contrast, targeting pas‐4 suppressed multiplication of in vitro‐dosed R. similis on carrot discs by 89 ± 2%. The suppression of transcript levels in R. similis recovered from carrot discs and the lack of suppression in P. coffeae suggest that the higher activity of R. similis and hence increased rate of feeding than P. coffeae (Stoffelen et al., 1999) results in uptake of sufficient dsRNA to maintain an RNAi effect while cultured on carrot discs. Despite high expression of the pas‐4 in body wall muscles and a large knock‐down following in vitro treatment with dsRNA, there was no effect on nematode locomotion. C. elegans‐fed bacteria expressing pas‐4 dsRNA had uncoordinated movement in a single experiment but not in all experiments of the study (Simmer et al., 2003). There was also no effect on locomotion in our experiments with plant‐parasitic nematodes or C. elegans treated with dsRNA in solution. This may indicate an effect on the locomotory or other musculature, such as those in the pharynx required for feeding, that only becomes evident following consistent and long‐term exposure when nematodes are cultured on transgenic roots.
The 376‐bp act‐4 dsRNA molecule had four regions with >19 nt sequence identity with the R. similis act‐4 transcript, including one in the truncated dsRNA region, and suppressed its locomotion and multiplication after oral dosing, during culture on carrot discs and when a parasite on transgenic hairy roots. The results for P. coffeae were somewhat different. No effect was detected with the truncated act‐4 dsRNA molecule. The locomotion of P. coffeae orally treated with the 376‐bp act‐4 dsRNA molecule was suppressed, but the small reduction in its act‐4 transcript was not statistically significant. Transgenic act‐4 line 6 also showed a phenotypic effect of significantly reduced multiplication without the accompanying small fall in message abundance reaching statistical significance in contrast to act‐4 line 5. Possibly, even a small reduction in transcript abundance has phenotypic effects on P. coffeae. The truncated 122‐bp molecule reduced P. coffeae multiplication on transgenic roots despite lacking comparatively low sequence identity to the target sequence. Possibly, the strength of base pairing of nucleotide seed match in positions 2 to 7–8 nucleotides of the antisense strand of the siRNA (Birmingham et al., 2007) is a stronger predictor of a potential effect than overall sequence identity between dsRNA molecule and transcript target sequence. This matching did occur between the P. coffeae sequence and potential act‐4_t seed regions. The efficacy of the truncated version of the act‐4 dsRNA with a comparatively low sequence similarity to the actin genes and the lack of an effect in C. elegans despite the presence of >19nt exact sequence matches are both of interest. They indicate the importance of experimental confirmation of in silico analyses given a higher sequence similarity of at least 88% has been suggested as required for an RNAi effect (Parrish et al., 2000).
The strategy of identifying single dsRNA molecules that target conserved sequences across nematode genera probably increased the chances of selecting genes with important biological roles and hence a significant RNAi knock‐down phenotype. Identification of targets for R. similis and M. incognita were greatly aided by genetic information for these nematodes (Abad et al., 2008; Bird et al., 2015; Jacob et al., 2009; Opperman et al., 2008). The recently published genome of P. coffeae (Burke et al., 2015) should also prove useful for future work. A constraint on target identification in this work was the lack of genetic information for H. multicinctus and, at the time of selecting targets, P. coffeae. Identification of homologous sequences for these species relied on amplification with primers designed to conserved regions in C. elegans, R. similis and M. incognita of the genes of interest. Additionally, a constraint on in vitro screening using carrot discs is the inability to culture H. multicinctus, as carrot is not a host for that nematode, or M. incognita, which cannot be cultured on carrot discs. Another issue, discussed above, is an inability to ensure consistent exposure to the dsRNA. As such, this screen is likely to underestimate the effectiveness of an RNAi effect as compared to the transgenic root screen.
The availability of complete genetic sequences not only aids the identification of RNAi targets but also allows the assessment of off‐target potential. The transformed dsRNA sequences had no 21‐nt matches for carrot in GenBank or the published banana genomes (D'Hont et al., 2012; Davey et al., 2013). The dsCheck software was used to reduce off‐target potential of the dsRNAs designed for this work. It provides an accelerated off‐target search algorithm for siRNAs to identify exact and near nucleotide matches to the five representative genomes of Oryza sativa (Rice), Arabidopsis thaliana, Rattus norvegicus (Rat), Drosophila melanogaster and C. elegans (Naito et al., 2005). There are also no exact 21‐nt matches of the transformed dsRNA sequences to the ESTs available in GenBank. In silico sequence analysis provides a useful means of selecting molecules for further study and supports rational design to reduce likely risk. However, the potential hazard of transitive silencing along the target sequences and production of secondary siRNAs that could possible cause off‐target effects must be evaluated. It requires transgenic lines of the intended crop, in this case banana, because the risk of off‐target effects is heavily dependent on the context of RNAi molecules. Consequently, the assessment off‐target effects of RNAi molecules is a future research need as hairy roots of carrot cannot indicate the likelihood of such effects in other plants.
There is an inherent advantage to any approach that can replace the low specificity and high toxicity of current nematode control chemicals. There is, however, a possibility of an RNAi effect on a wide range of nontarget organisms due to the high conservation of these two target transcripts that requires consideration in the design of dsRNA molecules. Previous studies have concentrated on parasitism genes to enhance the safety of the approach either by design (Huang et al., 2006) or through a process of excluding genes with homology in nontarget organisms (Danchin et al., 2013). The disadvantage of this approach is that target genes will likely be too specific for a broad control of crop pests. A more fine‐grained approach provides a larger pool of potential targets from which to choose. It bases selection on the homology of a targeted gene region, as opposed to the homology of the entire gene. This increases the potential for the control of a range of nematodes of several genera with different modes of parasitism, as demonstrated in this work, and ensures a low homology to transcripts in nontarget organisms, including humans. A lack of food hazard is also suggested by a wide range of evidence that dsRNA is not a hazard in the human diet (Petrick et al., 2013). We consider this provides a prima facie case for the food safety of the antinematode RNAi defences. Several steps in risk assessment are essential for both food and environmental safety once transgenic events of interest are identified. A key need is banana plants and fruit for the recommended context‐specific safety evaluation in planta of a transgenic event (Arpaia et al., 2017). That approach is required by regulatory authorities for potential nontarget effects for each transformation event generated (e.g. EFSA, 2010). One appropriate approach is the comparative safety assessment paradigm as employed for already commercialized transgenic crops (Parrott et al., 2010; Petrick et al., 2013). Biosafety evaluations are planned in both glasshouses and the field for crop plant growth, agronomic characteristics and environmental impact on nontarget organisms selected by a range of criteria (Ahmad et al., 2016), as well as food safety assessments on fruit.
Environmental monitoring should be used to complement the preventative design strategy, particularly where genetic information for at‐risk organisms is lacking. The emphasis of environmental risk assessments often focuses on beneficial organisms (Romeis et al., 2011), particularly those related to the target pest. Therefore, beneficial nematodes in the soil around the crop need to be considered when plant‐parasitic species are targeted for RNAi. Caenorhabditis and Cephalobus were selected for this work as both readily took up fluorescent molecules in vitro, unlike other free‐living nematodes tested. Both genera occurred in soil from a banana field in Uganda. Soaking C. elegans in dsRNA molecules designed to target its own act‐4 and pas‐4 genes mirrored the response of the parasitic nematode with suppression of transcript of both genes and a reduction in locomotion of act‐4 dsRNA‐treated nematodes. Transcript abundance and locomotion in C. elegans were not affected by the sequences targeted at the banana nematodes which have 85% identity for act‐4_t and 68% identity for pas‐4 to the C. elegans sequences. They may have provided too few exo‐siRNAs to trigger sufficient production by C. elegans of the secondary siRNAs that are required to amplify the signal (Billi et al., 2014). Locomotion of Cephalobus, a slow‐moving nematode, was not adversely affected by any of the dsRNAs. Previous work suggests many genera of free‐living soil nematodes are less readily affected by dsRNA than C. elegans (Wheeler et al., 2012). Sentinel species are a well‐established approach to indicate potential hazards (van der Schalie et al., 1999). Our results suggest C. elegans is appropriately susceptible for this use in monitoring RNAi‐mediated hazards to nontarget nematodes.
These results establish a framework for selecting gene targets for RNAi‐based control of crop pests that are from several distinct genera. This work concentrates on nematode pests of banana, but the approach is equally applicable to any group of pests and pathogens for which common and effective target sequences can be identified. Genetic resources and screens for efficacy are available for the approach to be extended to insects (Joga et al., 2016), viruses (Galvez et al., 2014) and fungi (Salame et al., 2011).
Experimental procedures
Identifying sequences of interest
Genes of interest were identified by searches of the GenBank database (http://www.ncbi.nlm.nih.gov; Benson et al., 2013), the NEMBASE4 EST database (http://www.nematodes.org/nembase4/; Elsworth et al., 2011), the nematode.net EST database (http://www.nematode.net/; Martin et al., 2015) and Wormbase (http://www.wormbase.org/ release WS227; Howe et al., 2016). The search was for gene EST sequences present in more than one of R. similis, Pratylenchus spp., H. multicinctus and Meloidogyne spp. and with a severe RNAi phenotype in Caenorhabditis elegans. Alignments using Clustal Omega version 1.2.3 (http://www.ebi.ac.uk/Tools/msa/clustalo/; Sievers et al., 2011) of homologous gene sequences were used to identify regions for primer design with high identity to ensure amplification from all targeted species. These primers were designed using Primer3Plus (http://primer3plus.com/cgi-bin/dev/primer3plus.cgi; Untergasser et al., 2012) and used to amplify gene sequences by standard PCR from nematodes where a database sequence was not available. Sequences were cloned into pCR‐Blunt II‐TOPO (Invitrogen, Carlsbad, CA) and sequenced from the M13 forward and reverse primer sites on that plasmid.
Generating dsRNA expressing constructs
DNA fragments of the target genes were amplified by PCR using specific primers with restriction site linkers (Table S1) to generate expression vectors. A modified GFP sequence (Haseloff et al., 1997) was used in to provide the nontarget control treatment. For in vitro experiments, the DNA fragments were subcloned into the TA‐cloning vector pCR‐Blunt II‐TOPO (Invitrogen) and then cloned into the NheI site of the vector L4440 (pPD129.36; Timmons and Fire, 1998). The resultant constructs were verified by DNA sequencing.
Hairpin vectors were generated with the same DNA fragments. They were cloned in a sense and antisense orientation either side of the spacer (loop) intron in the pHANNIBAL vector (Wesley et al., 2001). The act‐4_t fragment was cloned 5′–3′ into the XhoI and KpnI sites and 3′–5′ into the XbaI and HindIII sites of the vector. The pas‐4 fragment was cloned 5′–3′ into the EcoRI and KpnI sites and 3′–5′ into the XbaI and HindIII sites of the vector. The hairpin cassette containing, in 5′–3′ order, the CaMV35S promoter, the sense orientation target fragment, the spacer intron, the antisense orientation target fragment and an OCS 3′ region was then ligated into the NotI site of the pART27 binary vector (Gleave, 1992). Vector sequences were checked by sequencing then transformed into Agrobacterium rhizogenes strain R1000 (Chilton et al., 1982) by the freeze–thaw method.
Nematode culture
Radopholus similis and P. coffeae were maintained on carrot disc cultures and were collected by washing the carrot discs in Petri dishes with 15 mL tap water overnight. The water was transferred to 15 mL tubes and the nematodes allowed to settle for 1 h before transfer to a 1.5‐mL microcentrifuge tube using a glass pipette. H. multicinctus and M. incognita were maintained on banana plant roots and collected from chopped banana roots using a misting chamber (Southey, 1986) into 50‐mL tubes that were changed every 24 h for 3 days. Nematodes collected at the bottom of the tube were transferred to 1.5‐mL microcentrifuge tubes with a glass pipette.
The bacterial‐feeding nematode Cephalobus sp. wild isolate (strain DWF1301) was provided by CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Cephalobus sp. and C. elegans nematodes were maintained on lawns of Escherichia coli strain OP50 on Nematode Growth Medium (NGM) agar plates (Dusenbery et al., 1975) and collected by washing plates with 2 mL sterile tap water for 15 min and collecting nematodes into 1.5‐mL microcentrifuge tubes.
Prior to all experiments, nematodes were surface‐sterilized with 1 mL of sterilization mix (0.1% kanamycin, 0.1% penicillin G, 0.1% streptomycin sulphate, 50 μg/mL amphotericin B, 0.1% cetyltrimethylammonium bromide [CTAB]) for 30 min followed by five washes with 1 mL sterile tap water.
In vitro uptake of dsRNA
Synthesis of complementary single‐stranded RNAs (ssRNAs) was from the T7 promoters located either side of the multiple cloning site (MCS) in each nematode targeting L4440 construct following independent digestion with KpnI and SacI. Synthesis of ssRNA, annealing to dsRNA and dsRNA purification were by Ambion Megascript T7 RNAi kit (Invitrogen) according to the instructions provided. Three replicates of 200 nematodes were soaked in 100 μg/mL dsRNA in M9 buffer with 100 mm Octopamine at 25 °C in the dark for 24 h in 1.5‐mL tubes. This neurotransmitter has been used before as described for both Pratylenchus and Meloidogyne without adverse effects (Rosso et al., 2005; Tan et al., 2013).
Locomotion assay
Nematodes treated with dsRNA or collected from carrot hairy roots were aliquoted onto water agar plates (1% agar, 0.25% HEPES, 0.25% Tween‐20, pH 7.2). Video capture of locomotion of >15 active nematodes per treatment over 30 s was recorded at four frames per second and 1.25× magnification using a Q Imaging Micropublisher 3.3 RTV camera attached to a Leica M165C stereomicroscope. Track lengths of the imaged nematodes were measured over at least 30 s on each occasion using the wrMTrck plugin (Nussbaum‐Krammer et al., 2015) for Image J (Schneider et al., 2012) version 1.46r and exported to an Excel worksheet for subsequent analysis.
Carrot disc culture
Sterile nematodes treated for 16 hr with 100 μg/mL dsRNA were transferred to sterile carrot discs on 1% agar, 10 replicates of 250 nematodes per culture, and incubated at 25 °C in the dark for 56 days. Every 7 days, 100 μg/mL dsRNA in 1 ml M9 buffer was applied to each carrot disc culture. Nematodes were collected by washing with tap water, counted and half were used for the locomotion assay and half flash‐frozen for quantitative RT‐PCR analysis.
Hairy root cultures
Hairy root cultures were established on Petri dishes of 1% agar modified White's medium (MW; Becard and Fortin, 1988) with 50 μg/mL kanamycin selection from transformed carrot discs as described by Cardarelli et al. (1987). Expression of hairpin constructs was confirmed by RT‐PCR using primers designed to amplify the dsRNA molecules with amplification of the carrot actin‐7‐like gene as an internal control (Table S2). Cultures with root extension rates of 10 ± 2 mm/week were selected to provide 10 transformation events per construct (gfp, act‐4_t and pas‐4). An initial screen to identify promising lines used three replicate hairy root cultures per transformation event, subsequent screens on the best lines were with 15 replicate cultures per transformation event. For nematode challenge, approximately 200 nematodes were inoculated onto the agar next to several growing root tips and incubated on the roots at 25 °C in the dark for 12 weeks. Mixed stage R. simlis and P. coffeae and M. incognita J2s were washed from the hairy root cultures using sterile tap water, counted and flash‐frozen for quantitative RT‐PCR analysis.
Quantitative RT‐PCR
RNA was extracted from collected nematode samples by SV Total RNA Isolation System (Promega, Fitchburg, WI), which included an on‐column DNase treatment. Synthesis of cDNA was from 1 μg total RNA with 200 units Superscript II Reverse Transcriptase, and Oligo (dT) primers (Invitrogen). A one in 10 dilution of cDNA (100 ng/reaction) was used as template for qPCR analysis with gene‐specific primers with efficiencies >90% and <110% (Table S2) added to Agilent Brilliant II SYBR Green Master Mix (Agilent, Santa Clara, CA, US). Cycling conditions were 5 min at 95 °C followed by 40 cycles of 95 °C for 10 s and 55 °C for 30 s. A dissociation curve analysis was carried out at the end of each qPCR experiment to monitor reactions for nonspecific amplification. Quantification was the mean of four technical replicates for each of three biological replicates per experiment and was normalized to cell division control protein 42 (cdc‐42) for R. similis, heat‐shock protein 90 (hsp‐90) for P. coffeae, 18S ribosomal RNA sequences for M. incognita and C. elegans. Experiments were carried out using an Mx3005p qPCR Cycler (Agilent) and analysed in MxPro qPCR software (Agilent).
Conflict of interest
The authors declare they have no conflict of interest.
Supporting information
Figure S1 Alignments of (a) act‐4 and (b) pas‐4 dsRNA molecules and their targets in the plant‐parasitic nematodes Radopholus similis, Pratylenchus coffeae, Helicotylenchus multicinctus and Meloidogyne incognita and the nontarget, Caenorhabditis elegans.
Table S1 Primers used for dsRNA construct cloning. Linker restriction sites used for cloning are indicated in bold italics.
Table S2 Primers used for transcript screening and quantification. Genes used as internal controls for normalising quantification indicated in bold.
Video S1 Movement of Radopholus similis and Pratylenchus coffeae across 1% agar plates following treatment with two dsRNA molecules targeting act‐4, a dsRNA molecule targeting pas‐4 or a non‐targeting gfp sequence. Each nematode is representative of the mean distance moved for that species and treatment (N>15). Scale bar = 500μm.
Acknowledgements
We thank United States Agency for International Development (USAID) for financial support as part of the Agricultural Biotechnology Support Project II (ABSPII) programme, the Biotechnology and Biological Sciences Research Council (BBSRC) and Department for International Development (DFID) for financial support as part of the Sustainable Agriculture Research for International Development (SARID; BB/F004001/1) programme.
References
- Abad, P. , Gouzy, J. , Aury, J.M. , Castagnone‐Sereno, P. , Danchin, E.G. , Deleury, E. , Perfus‐Barbeoch, L. et al (2008) Genome sequence of the metazoan plant‐parasitic nematode Meloidogyne incognita . Nat. Biotechnol. 26, 909–915. [DOI] [PubMed] [Google Scholar]
- Ahmad, A. , Negri, I. , Oliveira, W. , Brown, C. , Asiime, P. , Sammons, B. , Horak, M. et al (2016) Transportable data from non‐target arthropod field studies for the environmental risk assessment of genetically modified maize expressing an insecticidal double‐stranded RNA. Transgenic Res. 25, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia, S. , Birch, A.N.E. , Kiss, J. , van Loon, J.J. , Messéan, A. , Nuti, M. , Perry, J.N. et al (2017) Assessing environmental impacts of genetically modified plants on non‐target organisms: the relevance of in planta studies. Sci. Total Environ. 583, 123–132. [DOI] [PubMed] [Google Scholar]
- Auer, C. and Frederick, R. (2009) Crop improvement using small RNAs: applications and predictive ecological risk assessments. Trends Biotechnol. 27, 644–651. [DOI] [PubMed] [Google Scholar]
- Becard, G. and Fortin, J.A. (1988) Early events of vesicular‐arbuscular mycorrhiza formation on Ri T‐DNA transformed roots. New Phytol. 108, 211–218. [DOI] [PubMed] [Google Scholar]
- Benson, D.A. , Cavanaugh, M. , Clark, K. , Karsch‐Mizrachi, I. , Lipman, D.J. , Ostell, J. and Sayers, E.W. (2013) GenBank. Nucleic Acids Res. 41, D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi, A.C. , Fischer, S.E. and Kim, J.K. (2014) Endogenous RNAi pathways in C. elegans. In: WormBook, The Online Review of C. elegans Biology (The C. elegans Research Community, eds). Wormbook: http://www.wormbook.org, doi/10.1895/wormbook.1.170.1. [DOI] [PMC free article] [PubMed]
- Bird, D.M. , Williamson, V.M. and Opperman, C.H. (2015) Exploiting solved genomes of plant‐parasitic nematodes to understand parasitism. Adv. Bot. Res. 73, 241–258. [Google Scholar]
- Birmingham, A. , Anderson, E. , Sullivan, K. , Reynolds, A. , Boese, Q. , Leake, D. , Karpilow, J. et al (2007) A protocol for designing siRNAs with high functionality and specificity. Nat. Protoc. 2, 2068–2078. [DOI] [PubMed] [Google Scholar]
- Bridge, J. , Price, N.S. and Kofi, P. (1995) Plant parasitic nematodes of plantain and other crops in Cameroon, West Africa. Fundam. Appl. Nematol. 18, 251–260. [Google Scholar]
- Burke, M. , Scholl, E.H. , Bird, D.M. , Schaff, J.E. , Coleman, S. , Crowell, R. , Diener, S. et al (2015) The plant parasite Pratylenchus coffeae carries a minimal nematode genome. Nematology, 17, 621–637. [Google Scholar]
- Cardarelli, M. , Mariotti, D. , Pomponi, M. , Spanò, L. , Capone, I. and Costantino, P. (1987) Agrobacterium rhizogenes T‐DNA genes capable of inducing hairy root phenotype. Mol. Gen. Genet. 209, 475–480. [DOI] [PubMed] [Google Scholar]
- Chilton, M.‐D. , Tepfer, D.A. , Petit, A. , David, C. , Casse‐Delbart, F. and Tempé, J. (1982) Agrobacterium rhizogenes inserts T‐DNA into the genomes of the host plant root cells. Nature, 295, 432–434. [Google Scholar]
- Custodia, N. , Won, S.J. , Novillo, A. , Wieland, M. , Li, C. and Callard, I.P. (2001) Caenorhabditis elegans as an environmental monitor using DNA microarray analysis. Ann. N. Y. Acad. Sci. 948, 1749–6632. [DOI] [PubMed] [Google Scholar]
- Danchin, E.G.J. , Arguel, M.J. , Campan‐Fournier, A. , Perfus‐Barbeoch, L. , Magliano, M. , Rosso, M.‐N. , Da Rocha, M. et al (2013) Identification of novel target genes for safer and more specific control of root‐knot nematodes from a pan‐genome mining. PLoS Pathog. 9, e1003745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey, M.W. , Gudimella, R. , Harikrishna, J.A. , Sin, L.W. , Khalid, N. and Keulemans, J. (2013) A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter‐and intra‐specific Musa hybrids. BMC Genom. 14, 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Hont, A. , Denoeud, F. , Aury, J.M. , Baurens, F.C. , Carreel, F. , Garsmeur, O. , Noel, B. et al (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature, 488, 213–217. [DOI] [PubMed] [Google Scholar]
- Dusenbery, D.B. , Sheridan, R.E. and Russell, R.L. (1975) Chemotaxis‐defective mutants of the nematode Caenorhabditis elegans . Genetics, 80, 297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA . (2010) Guidance on the environmental risk assessment of genetically modified plants. EFSA J. 8, 1879. [Google Scholar]
- Elsworth, B. , Wasmuth, J. and Blaxter, M. (2011) NEMBASE4: the nematode transcriptome resource. Int. J. Parasitol. 41, 881–894. [DOI] [PubMed] [Google Scholar]
- FAOSTAT . (2014) Agriculture Data. www.fao.org/ag.
- Fire, A. , Xu, S.Q. , Montgomery, M.K. , Kostas, S.A. , Driver, S.E. and Mello, C.C. (1998) Potent and specific genetic interference by double‐stranded RNA in Caenorhabditis elegans . Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Frisson, E. and Sharrock, S. (1999) The economic, social and nutritional importance of banana in the world In Proceedings of International Symposium on Bananas and Food Security (Picq C., Foure E. and Frisson E.A., eds), pp. 21–35. Montpellier, France: IPGRI. [Google Scholar]
- Galvez, L.C. , Banerjee, J. , Pinar, H. and Mitra, A. (2014) Engineered plant virus resistance. Plant Sci. 228, 11–25. [DOI] [PubMed] [Google Scholar]
- Gleave, A.P. (1992) A versatile binary vector system with a T‐DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207. [DOI] [PubMed] [Google Scholar]
- Gowen, S.C. , Quénéherve, P. and Fogain, R. (2005) Nematode parasites of bananas and plantains In Plant Parasitic Nematodes in Subtropical and Tropical Agriculture, 2nd ed. (Luc M., Sikora R.A. and Bridge J., eds), pp. 611–643. Wallingford, UK: CABI. [Google Scholar]
- Haseloff, J. , Siemering, K.R. , Prasher, D.C. and Hodge, S. (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl Acad. Sci. USA, 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser, S. and Amougou, D. (2010) Plantain (Musa spp.) Cropping systems of Southern Cameroon. Acta Hortic. 879, 495–508. [Google Scholar]
- Heinemann, J.A. , Agapito‐Tenfen, S.Z. and Carman, J.A. (2013) A comparative evaluation of the regulation of GM crops or products containing dsRNA and suggested improvements to risk assessments. Environ. Int. 55, 43–55. [DOI] [PubMed] [Google Scholar]
- Howe, K.L. , Bolt, B.J. , Cain, S. , Chan, J.C. , Chen, W.J. , Davis, P. , Done, J. et al (2016) WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 44, D774–D780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, G.Z. , Allen, R. , Davis, E.L. , Baum, T.J. and Hussey, R.S. (2006) Engineering broad root‐knot resistance in transgenic plants by RNAi silencing of a conserved and essential root‐knot nematode parasitism gene. Proc. Natl Acad. Sci. USA, 103, 14302–14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, H.M.M. , Alkharouf, N.W. , Meyer, S.L.F. , Aly, M.A.M. , Gamal El‐Din, A.Y. , Hussein, E.H.A. and Matthews, B.F. (2011) Post‐transcriptional gene silencing of root‐knot nematode in transformed soybean roots. Exp. Parasitol. 127, 90–99. [DOI] [PubMed] [Google Scholar]
- Jacob, J.E.M. , Vanholme, B. , Van Leeuwen, T. and Gheysen, G. (2009) A unique genetic code change in the mitochondrial genome of the parasitic nematode Radopholus similis . BMC Res. Notes, 2, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joga, M.R. , Zotti, M.J. , Smagghe, G. and Christiaens, O. (2016) RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front. Physiol. 7, 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, S. , Gheysen, G. and Subramaniam, K. (2012) RNA interference in Pratylenchus coffeae: knock down of Pc‐pat‐10 and Pc‐unc‐87 impedes migration. Mol. Biochem. Parasitol. 186, 51–59. [DOI] [PubMed] [Google Scholar]
- Kamola, P.J. , Nakano, Y. , Takahashi, T. , Wilson, P.A. and Ui‐Tei, K. (2015) The siRNA non‐seed region and its target sequences are auxiliary determinants of off‐target effects. PLoS Comput. Biol. 11, e1004656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashaija, I.N. , Speijer, P.R. , Gold, C.S. and Gowen, S.C. (1994) Occurrence, distribution and abundance of plant parasitic nematodes of bananas in Uganda. Afr. Crop Sci. J. 2, 99–104. [Google Scholar]
- Kaur, A. , Kumar, A. and Reddy, M.S. (2016) RNA Interference (RNAi) and its role in crop improvement: a review In Plant Tissue Culture: Propagation, Conservation and Crop Improvement (Anis M. and Ahmad N., eds), pp. 379–394. Singapore: Springer. [Google Scholar]
- Klink, V.P. , Kim, K.H. , Martins, V. , MacDonald, M.H. , Beard, H.S. , Alkharouf, N.W. , Lee, S.K. et al (2009) A correlation between host‐mediated expression of parasite genes as tandem inverted repeats and abrogation of development of female Heterodera glycines cyst formation during infection of Glycine max . Planta, 230, 53–71. [DOI] [PubMed] [Google Scholar]
- Li, J. , Todd, T.C. , Oakley, T.R. , Lee, J. and Trick, H.N. (2010) Host‐derived suppression of nematode reproductive and fitness genes decreases fecundity of Heterodera glycines Ichinohe. Planta, 232, 775–785. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Wang, K. and Xie, H. (2015a) Cathepsin B cysteine proteinase is essential for the development and pathogenesis of the plant parasitic nematode Radopholus similis . Int. J. Biol. Sci. 11, 1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Wang, K. , Xie, H. , Wang, Y.T. and Wang, D.W. (2015b) A nematode calreticulin, Rs‐CRT, is a key effector in reproduction and pathogenicity of Radopholus similis . PLoS ONE, 10, e0129351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley, C.J. , Davies, L.J. and Urwin, P.E. (2012) RNA interference in plant parasitic nematodes: a summary of the current status. Parasitology, 139, 630–640. [DOI] [PubMed] [Google Scholar]
- Lorenzen, J. , Tenkouano, A. , Bandyopadhyay, R. , Vroh, B. , Coyne, D. and Tripathi, L. (2010) Overview of banana and plantain (Musa spp.) improvement in Africa: past and future. Acta Hortic. 879, 595–603. [Google Scholar]
- Martin, J. , Rosa, B.A. , Ozersky, P. , Hallsworth‐Pepin, K. , Zhang, X. , Bhonagiri‐Palsikar, V. , Tyagi, R. et al (2015) Helminth.net: expansions to Nematode.net and an introduction to Trematode.net. Nucleic Acids Res. 43, D698–D706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito, Y. , Yamada, T. , Matsumiya, T. , Ui‐Tei, K. , Saigo, K. and Morishita, S. (2005) dsCheck: highly sensitive off‐target search software for dsRNA‐mediated RNA interference. Nucleic Acids Res. 33, W589–W591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgrove, L. and Hauser, S. (2014) Improving plantain (Musa spp. AAB) yields on smallholder farms in West and Central Africa. Food Secur. 6, 501–514. [Google Scholar]
- Nussbaum‐Krammer, C.I. , Neto, M.F. , Brielmann, R.M. , Pedersen, J.S. and Morimoto, R.I. (2015) Investigating the spreading and toxicity of prion‐like proteins using the metazoan model organism C. elegans . J. Vis. Exp. 95, e52321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman, C.H. , Bird, D.M. , Williamson, V.M. , Rokhsar, D.S. , Burke, M. , Cohn, J. , Cromer, J. et al (2008) Sequence and genetic map of Meloidogyne hapla: a compact nematode genome for plant parasitism. Proc. Natl Acad. Sci. USA, 105, 14802–14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, R. and Vuylsteke, D. (1996) Improving plantain and banana‐based systems In Plantain and Banana Production and Research in West and Central Africa (Ortiz R. and Akoroda M.O., eds), pp. 2–7. Ibadan, Nigeria: IITA. [Google Scholar]
- Papaevgeniou, N. and Chondrogianni, N. (2014) The ubiquitin proteasome system in Caenorhabditis elegans and its regulation. Redox. Biol. 2, 333–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish, S. , Fleenor, J. , Xu, S. , Mello, C. and Fire, A. (2000) Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol. Cell, 6, 1077–1087. [DOI] [PubMed] [Google Scholar]
- Parrott, W. , Chassy, B. , Ligon, J. , Meyer, L. , Petrick, J. , Zhou, J. , Herman, R. et al (2010) Application of food and feed safety assessment principles to evaluate transgenic approaches to gene modulation in crops. Food Chem. Toxicol. 48, 1773–1790. [DOI] [PubMed] [Google Scholar]
- Patel, N. , Hamamouch, N. , Li, C.Y. , Hussey, R.S. , Mitchum, M. , Baum, T. , Wang, X. et al (2008) Similarity and functional analyses of expressed parasitism genes in Heterodera schachtii and Heterodera glycines . J. Nematol. 40, 299–310. [Google Scholar]
- Patel, N. , Hamamouch, N. , Li, C.Y. , Hewezi, T. , Hussey, R.S. , Baum, T. , Mitchum, M. et al (2010) A nematode effector protein similar to annexins in host plants. J. Exp. Bot. 61, 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrick, J.S. , Brower‐Toland, B. , Jackson, A.L. and Kier, L.D. (2013) Safety assessment of food and feed from biotechnology‐derived crops employing RNA‐mediated gene regulation to achieve desired traits: a scientific review. Regul. Toxicol. Pharmacol. 66, 167–176. [DOI] [PubMed] [Google Scholar]
- Roberts, A.F. , Devos, Y. , Lemgo, G.N.Y. and Zhou, X. (2015) Biosafety research for non‐target organism risk assessment of RNAi‐based GE plants. Front. Plant. Sci. 6, 958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderick, H. , Mbiru, E. , Coyne, D. , Tripathi, L. and Atkinson, H.J. (2012) Quantitative digital imaging of banana growth suppression by plant parasitic nematodes. PLoS ONE, 7, e53355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis, J. , Hellmich, R.L. , Candolfi, M.P. , Carstens, K. , De Schrijver, A. , Gatehouse, A.M. , Herman, R.A. et al (2011) Recommendations for the design of laboratory studies on non‐target arthropods for risk assessment of genetically engineered plants. Transgenic Res. 20, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.N. , Dubrana, M.P. , Cimbolini, N. , Jaubert, S. and Abad, P. (2005) Application of RNA interference to root‐knot nematode genes encoding esophageal gland proteins. MPMI, 18, 615–620. [DOI] [PubMed] [Google Scholar]
- Rual, J.F. , Klitgord, N. and Achaz, G. (2007) Novel insights into RNAi off‐target effects using C. elegans paralogs. BMC Genom. 8, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salame, T.M. , Ziv, C. , Hadar, Y. and Yarden, O. (2011) RNAi as a potential tool for biotechnological applications in fungi. Appl. Microbiol. Biotechnol. 89, 501–512. [DOI] [PubMed] [Google Scholar]
- van der Schalie, W.H. , Gardner, H.S. Jr. , Bantle, J.A. , De Rosa, C.T. , Finch, R.A. , Reif, J.S. , Reuter, R.H. et al (1999) Animals as sentinels of human health hazards of environmental chemicals. Environ. Health Perspect. 107, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C.A. , Rasband, W.S. and Eliceiri, K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, F. , Wilm, A. , Dineen, D. , Gibson, T.J. , Karplus, K. , Li, W. , Lopez, R. et al (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer, F. , Moorman, C. , van der Linden, A.M. , Kuijk, E. , van den Berghe, P.V.E. , Kamath, R.S. , Fraser, A.G. et al (2003) Genome‐wide RNAi of C. elegans using the hypersensitive rrf‐3 strain reveals novel gene functions. PLoS Biol. 1, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhu, A.S. , Maier, T.R. , Mitchum, M.G. , Hussey, R.S. , Davis, E.L. and Baum, T.J. (2009) Effective and specific in planta RNAi in cyst nematodes: expression interference of four parasitism genes reduces parasitic success. J. Exp. Bot. 60, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southey, J. (1986) Laboratory Methods for Work with Plant and Soil Nematodes. Reference Book 402. London: Her Majesty's Stationery Office. [Google Scholar]
- Ssango, F. , Speijer, P.R. , Coyne, D.L. and De Waele, D. (2004) Path analysis: a novel approach to determine the contribution of nematode damage to East African Highland banana (Musa spp., AAA) yield loss under two crop management practices in Uganda. Field Crops Res. 90, 177–187. [Google Scholar]
- Stoffelen, R. , Jimenez, M.I. , Dierckxsens, C. , Tam, V.T.T. , Swennen, R. and de Waele, D. (1999) Effect of time and inoculum density on the reproductive fitness of Pratylenchus coffeae and Radopholus similis populations on carrot disks. Nematology, 1, 243–250. [Google Scholar]
- Tan, J.A.C.H. , Jones, M.G.K. and Fosu‐Nyarko, J. (2013) Gene silencing in root lesion nematodes (Pratylenchus spp.) significantly reduces reproduction in a plant host. Exp. Parasitol. 133, 166–178. [DOI] [PubMed] [Google Scholar]
- Timmons, L. and Fire, A. (1998) Specific interference by ingested dsRNA. Nature, 395, 854. [DOI] [PubMed] [Google Scholar]
- Untergasser, A. , Cutcutache, I. , Koressaar, T. , Ye, J. , Faircloth, B.C. , Remm, M. and Rozen, S.G. (2012) Primer3–new capabilities and interfaces. Nucleic Acids Res. 40, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwin, P.E. , Lilley, C.J. and Atkinson, H.J. (2002) Ingestion of double‐stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant Microbe Interact. 15, 747–752. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V. , Helliwell, C.A. , Smith, N.A. , Wang, M.B. , Rouse, D.T. , Liu, Q. , Gooding, P.S. et al (2001) Construct design for efficient, effective and high‐throughput gene silencing in plants. Plant J. 27, 581–590. [DOI] [PubMed] [Google Scholar]
- West, P.C. , Gerber, J.S. , Engstrom, P.M. , Mueller, N.D. , Brauman, K.A. , Carlson, K.M. , Cassidy, E.S. et al (2014) Leverage points for improving global food security and the environment. Science, 345, 325–328. [DOI] [PubMed] [Google Scholar]
- Wheeler, D. , Darby, B.J. , Todd, T.C. and Herman, M.A. (2012) Several grassland soil nematode species are insensitive to RNA‐mediated interference. J. Nematol. 44, 92–101. [PMC free article] [PubMed] [Google Scholar]
- Winston, W.M. , Sutherlin, M. , Wright, A.J. , Feinberg, E.H. and Hunter, C.P. (2007) Caenorhabditis elegans SID‐2 is required for environmental RNA interference. Proc. Natl Acad. Sci. USA, 104, 10565–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, B.C. , Veluthambi, K. and Subramaniam, K. (2006) Host‐generated double stranded RNA induces RNAi in plant‐parasitic nematodes and protects the host from infection. Mol. Biochem. Parasitol. 148, 219–222. [DOI] [PubMed] [Google Scholar]
- Yamada, T. and Morishita, S. (2005) Accelerated off‐target search algorithm for siRNA. Bioinformatics, 21, 1316–1324. [DOI] [PubMed] [Google Scholar]
- Yang, J. and Han, Z.J. (2013) Optimisation of RNA interference‐mediated gene silencing in Helicoverpa armigera . Aust. Entomol. 53, 83–88. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Alignments of (a) act‐4 and (b) pas‐4 dsRNA molecules and their targets in the plant‐parasitic nematodes Radopholus similis, Pratylenchus coffeae, Helicotylenchus multicinctus and Meloidogyne incognita and the nontarget, Caenorhabditis elegans.
Table S1 Primers used for dsRNA construct cloning. Linker restriction sites used for cloning are indicated in bold italics.
Table S2 Primers used for transcript screening and quantification. Genes used as internal controls for normalising quantification indicated in bold.
Video S1 Movement of Radopholus similis and Pratylenchus coffeae across 1% agar plates following treatment with two dsRNA molecules targeting act‐4, a dsRNA molecule targeting pas‐4 or a non‐targeting gfp sequence. Each nematode is representative of the mean distance moved for that species and treatment (N>15). Scale bar = 500μm.


