Summary
Autophagy is a major and conserved pathway for delivering and recycling unwanted proteins or damaged organelles to be degraded in the vacuoles. AuTophaGy‐related (ATG) protein 18a has been established as one of the essential components for autophagy occurrence in Arabidopsis thaliana. We previously cloned the ATG18a homolog from Malus domestica (MdATG18a) and monitored its responsiveness to various abiotic stresses at the transcriptional level. However, it is still unclear what its function is under abiotic stress in apple. Here, we found that heterologous expression of MdATG18a in tomato plants markedly enhanced their tolerance to drought. Overexpression (OE) of that gene in apple plants improved their drought tolerance as well. Under drought conditions, the photosynthesis rate and antioxidant capacity were significantly elevated in OE lines when compared with the untransformed wild type (WT). Transcript levels of other important apple ATG genes were more strongly up‐regulated in transgenic MdATG18a OE lines than in the WT. The percentage of insoluble protein in proportion to total protein was lower and less oxidized protein accumulated in the OE lines than in the WT under drought stress. This was probably due to more autophagosomes being formed in the former. These results demonstrate that overexpression of MdATG18a in apple plants enhances their tolerance to drought stress, probably because of greater autophagosome production and a higher frequency of autophagy. Those processes help degrade protein aggregation and limit the oxidation damage, thereby suggesting that autophagy plays important roles in the drought response.
Keywords: autophagy, apple, MdATG18a, drought, reactive oxygen species, oxidized protein
Introduction
Plants are inevitably challenged by various environmental stresses such as drought, salt, intense irradiance and heat. Because climates are warming and water supplies are becoming limited, drought stress is now a global phenomenon that significantly threatens future crop production (Zhao and Running, 2010). Drought conditions generate and accumulate reactive oxygen species (ROS), leading to membrane disruption, enzyme dysfunction and protein oxidation and aggregation (Tsugane et al., 1999). In plant cells, ROS can be scavenged through both enzymatic and nonenzymatic pathways (Apel and Hirt, 2004). The enzymatic detoxification system involves crucial enzymes such as superoxide dismutase (SOD), catalase (CAT), peroxidase (POD), ascorbate peroxidase (APX), dehydroascorbate reductase (DHAR), monodehydroascorbate reductase (MDHAR) and glutathione peroxidase (GR). While the nonenzymatic scavengers include efficient antioxidants such as ascorbate and glutathione (Foyer and Noctor, 2009), these ROS‐scavenging systems are crucial to the plant tolerance under stresses.
Autophagy, a conserved cellular process in eukaryotes, plays an important role in recycling nutrients and cytoplasmic components, as well as in conferring tolerance to biotic and abiotic stresses (Han et al., 2015; Havé et al., 2016; Mizushima and Komatsu, 2011; Ryabovol and Minibayeva, 2016; Wang et al., 2016, 2015b; Xiong et al., 2005). In the cells of plants that undergo starvation or other stresses, production of double‐membrane structures, the autophagosomes, is induced. Aggregated proteins or damaged organelles in the cytoplasm are engulfed by the autophagosomes and are degraded for recycle when autophagosomes fuse with the vacuole. Plant autophagy can be activated by various abiotic factors, for example, nutrient starvation, oxidative stress, salinity, drought, heat and hypoxia. These stresses enhance the accumulation of ROS and oxidized proteins. Autophagy‐defective mutants are hypersensitive to those stress conditions (Chen et al., 2015; Han et al., 2011; Liu et al., 2009; Xiong et al., 2005, 2007b; Zhou et al., 2014b). With reduced autophagic degradation rate, Arabidopsis RNAi‐AtATG18a plants are hypersensitive to hydrogen peroxide (H2O2) and accumulate more oxidized proteins than WT (Xiong et al., 2007b), indicating that autophagy is important in removing oxidized proteins under oxidative stress. Meanwhile, RNAi‐AtATG18a seedlings showed more severe growth inhibition than WT upon drought and salt treatment (Liu et al., 2009), implying autophagy as crucial participation in stress response. The Arabidopsis autophagy‐defective mutants, like atg5 and atg7, are demonstrated to have compromised tolerance to oxidative stress (Zhou et al., 2013). Moreover, rice Osatg10b mutants are sensitive to methyl viologen (MV) with high levels of oxidized proteins (Shin et al., 2009). Recent study in tomato found that overexpression of a heat‐shock transcription factor A1a (HsfA1a) confers drought tolerance by up‐regulating ATGs expression and inducing autophagosomes formation, which is achieved through the direct binding of HsfA1a to the promoters of ATG10 and ATG18f. These also suggest that autophagy functions in promoting plant survival under a water deficit (Wang et al., 2015b).
In yeast, the WD40 repeat‐containing protein Atg18 is able to bind phosphatidylinositol 3‐phosphate [PtdIns(3)P] and phosphatidylinositol 3,5‐bisphosphate [PtdIns(3,5)P2] (Krick et al., 2006). The PtdIns(3)P binding capacity of Atg18 is needed for efficient recruitment of Atg8 and Atg16 during phagophore formation at the phagophore assembly site (PAS) (Nair et al., 2010). The presence of Atg18/21 complex blocks access of Atg4 to Atg8‐phosphatidylethanolamine (PE) at PAS, preventing a premature cleavage event. However, following completion of the autophagosome, probably under regulation of Atg1, the Atg18/21 complex will dissociate and allow Atg4 to cleave Atg8 from PE. Therefore, the key aspect of post‐translational regulation of autophagy by Atg4 is closely related with Atg18/21 (Nair et al., 2010). Yeast Atg18 gene is required for starvation‐induced autophagy, as atg18 mutant is unable to accumulate autophagosomes in response to starvation (Barth et al., 2001; Guan et al., 2001). Arabidopsis has eight ATG18s homologs (a‐h), and each member shows a different transcript pattern. Only AtATG18a showed a significant increase in transcript level in both sucrose and nitrogen starvation conditions, as well as during artificial senescence and oxidative stress treatment (Xiong et al., 2005, 2007a). As stated earlier, RNAi‐AtATG18a plants, with defect in autophagosomes formation, are sensitive to oxidative stress, high salt, drought and necrotrophic pathogens (Lai et al., 2011; Liu and Bassham, 2012; Liu et al., 2009; Xiong et al., 2005). On the account of importance of ATG18a in autophagosomes formation and roles in stress response, we carried out our study on autophagy in apple plants. MdATG18a is the first autophagy gene cloned from apple (Malus domestica), and little is known about the function of autophagy in apple plants towards environmental stresses. We found that MdATG18a has conserved WD40 domains and is transcriptionally induced by various abiotic stresses including drought (Wang et al., 2014). As one of the most economically important fruits in the world, apple production faces great challenge towards drought stress in Loess Plateau, one of the main apple production regions in China. Therefore, identifying the genes responsible for drought tolerance and manipulating their expression in genetically modified crops is becoming a critical focus of molecular breeding programmes. To further investigate the function of MdATG18a and explore the initial role of autophagy in apple plants upon drought stress, we generated transgenic tomato and apple plants that overexpressed MdATG18a (OE lines) under the control of the CaMV 35S promoter. MdATG18a overexpression was associated with enhanced drought tolerance in both species. Under drought stress, when compared with WT, the MdATG18a transgenic apple plants showed higher increased expression of ATG genes and more autophagosomes. ROS‐scavenging systems were investigated, and we found higher enzymatic activities and more nonenzymatic antioxidants in transgenic apple plants than WT. Activities of CAT and POD, as well as enzymes in ascorbate–glutathione cycle, increased more greatly in transgenic apple plants than WT. The enhanced antioxidative activities and autophagosomes formation might have contributed to less oxidative damage and less accumulations of oxidized proteins observed in the apple transgenics. Thus, MdATG18a overexpression enhanced drought tolerance by increasing autophagy, which is thought to be involved in the degradation of oxidized proteins and the regulation of ROS levels under drought stress.
Results
Overexpression of MdATG18a in tomato enhances drought tolerance
Because MdATG18a expression is induced by drought (Wang et al., 2014), we overexpressed this gene for further analysis of its biological function under such stress. After its coding region was introduced into a plant‐overexpressing vector under the control of the CaMV35S promoter, we obtained nine transgenic lines (Figure S1a), and two transgenic tomato lines (OE‐1 and OE‐9) had single‐copy insertions (Figure S1b). These two lines constitutively overexpress MdATG18a with high mRNA transcript levels (Figure 1a). Under well‐watered conditions, performance did not differ between the WT and the transgenic lines (Figure 1b). However, when drought treatment was applied (i.e. irrigation withheld for 21 days), leaves from WT plants exhibited extensive symptoms of dehydration compared with only slight wilting in OE‐1 and OE‐9 lines after 14 days (Figure 1b). By Day 21, most of the WT leaves were necrotic and kraurotic while those of the transgenic lines remained green and vigorous.
Figure 1.
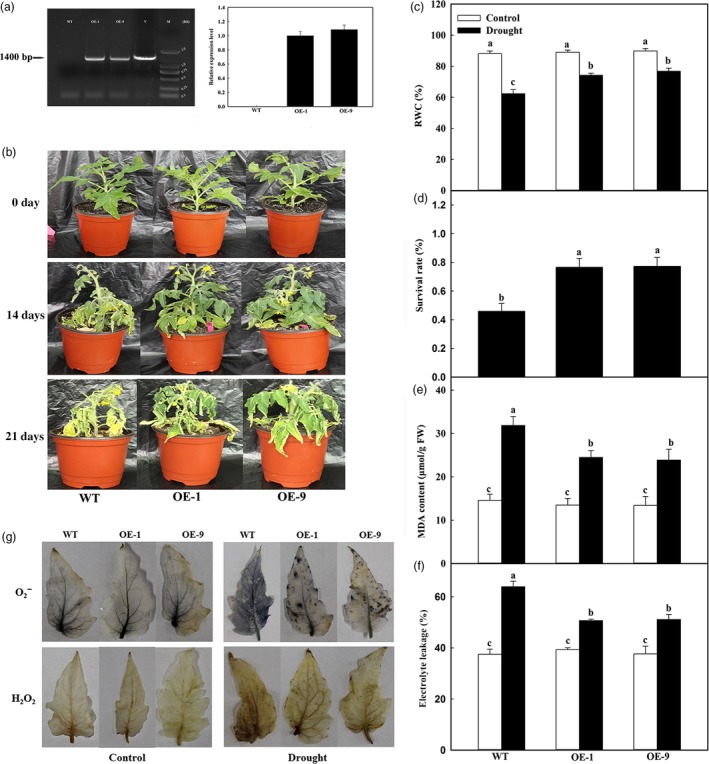
Drought stress tolerance and accumulations of H2O2 and O2 ‐ in MdATG18a‐overexpressing tomato. Water was withheld from 3‐week‐old plants for up to 21 days, followed by 3 days of recovery (rewatering). (a) PCR confirmation for transgenic tomato plants. Left panel: PCR with DNA; lanes: M, molecular marker DL2000; V, positive vector containing pCambia2300‐MdATG18a plasmid; WT, nontransformed wild type; OE‐1 and OE‐9, MdATG18a‐transgenic tomato lines. Right panel: Quantitative RT–PCR analysis of MdATG18a expression in leaves of WT and transgenic lines OE‐1 and OE‐9. (b) Increased tolerance in MdATG18a OE plants. (c) Comparisons of RWC among WT and OE lines after 21 days of drought treatment. (d) Survival rates of WT and transgenic plants at end of 3‐days recovery period. (e) MDA concentrations in WT and transgenic plants on Day 21 of drought treatment. (f) Electrolyte leakage in WT and transgenic plants on Day 21 of treatment. (g) Results from staining to detect H2O2 and O2 ‐ in leaves from glasshouse‐grown plants exposed to drought treatment for 21 days. Measurements of electrolyte leakage, MDA and RWC were made immediately after the tissues were collected. For H2O2 and , leaves were excised from plants on Day 21 of treatment and immediately placed in NBT for 4 h (H2O2 test) or DAB for 12 h ( test). Data are means of three replicates with SD. Different letters indicate significant differences between treatments, according to one‐way ANOVA Tukey's multiple range tests (P < 0.05).
Drought damage was evaluated by measuring relative water content (RWC), electrolyte leakage and levels of malondialdehyde (MDA), which are typical parameters used for assessing tolerance to abiotic stresses in crop plants (Levine et al., 1994; Pompelli et al., 2010; Tang et al., 2013; Wang et al., 2012b). Under well‐watered conditions, values for each of these parameters did not differ significantly among genotypes (Figure 1c–e). However, after 14 days of water deprivation, the RWCs of lines OE‐1 and OE‐9 were 19.03% and 23.07% higher, respectively, than the level calculated for the WT (Figure 1c). Survival rates also differed for drought‐stressed plants, that is 76% for the transgenics versus 46% for the WT (Figure 1d). For all plant types, MDA concentrations were increased in response to drought treatment, but the degree of increment was lower in the transgenics (Figure 1e). Finally, our comparison of electrolyte leakage measurements showed that values were 79.2% (OE‐1) and 79.9% (OE‐9) of that determined for the WT (Figure 1f).
The drought‐induced accumulation of highly reactive, toxic ROS can lead to oxidative stress, ultimately damaging various cell components. We used histochemical staining to examine the potential link between MdATG18a expression and production of oxide molecules and found that levels of both and H2O2 were greatly accumulated in response to drought. However, those accumulations were not as high in the OE lines as they were in the WT (Figure 1g).
Overexpression of MdATG18a in apple enhances drought tolerance
To further investigate the role of MdATG18a in drought‐stressed apple, we generated two overexpressing lines with high levels of mRNA transcripts and found that MdATG18a transcripts were increased by 32 and 36 times in lines OE‐3 and OE‐11, respectively (Figure 2a,b). Southern blot showed that there were positive hybridization signals in both OE‐3 and OE‐11 plants with single‐copy insertions (Figure S1c).
Figure 2.
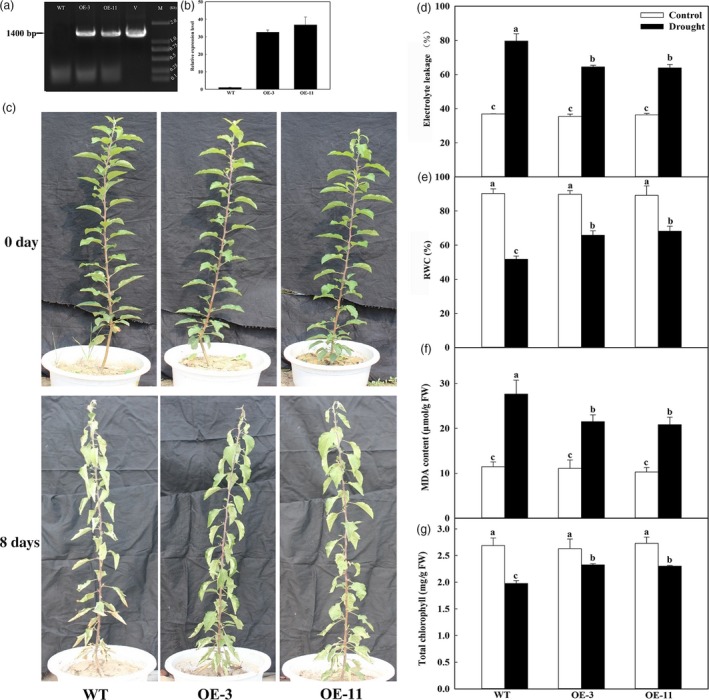
Drought tolerance by MdATG18a‐overexpressing apple. Water was withheld from 4‐month‐old plants for up to 8 days. (a) PCR with DNA; lanes: M, molecular marker DL2000; V, positive vector containing pCambia2300‐MdATG18a plasmid; WT, nontransformed wild‐type; OE‐3 and ‐11, MdATG18a‐transgenic lines. (b) qRT–PCR analysis of MdATG18a transcripts in lines OE‐3 and OE‐11; (c) drought tolerance in MdATG18a OE plants. (d) Electrolyte leakage in WT and transgenic plants after 8 days of drought stress. (e) Comparisons of RWC from WT and OE lines on Day 8 of treatment. (f) MDA concentrations in WT and transgenic plants on Day 8 of treatment. (g) Total chlorophyll concentrations in WT and transgenic plants on Day 8 of treatment. Data are means of three replicates with SD. Different letters indicate significant differences between treatments, according to one‐way ANOVA Tukey's multiple range tests (P < 0.05).
Under well‐watered control conditions, the phenotypes did not differ significantly between OE lines and the WT. However, after water was withheld for 8 days, the transgenics showed much less wilting and necrosis, and most of their leaves remained vigorous (Figure 2c). Furthermore, their electrolyte leakage was significantly lower than in WT plants on Day 8 (Figure 2d). Although RWCs for all plants were clearly decreased after drought treatment, the rate of that decline was slower in the transgenics, and values were 1.27–1.32 times that of WT plants (Figure 2e). Lower MDA concentrations were detected in the OE lines under drought treatment (Figure 2f). On Day 8 of treatment, total chlorophyll concentrations in OE‐3 and OE‐11 were, respectively, 1.16–1.18 times that of WT plants (Figure 2g). These data demonstrated that overexpression of MdATG18a results in less physiological damage to transgenic lines than to the WT under induced drought conditions.
Apple lines overexpressing MdATG18a maintain higher rates of photosynthesis under drought stress
The efficiency of photosynthesis is directly inhibited when stomata close in response to drought stress. To examine the tolerance phenotype of transgenic plants in this respect, we monitored their gas exchange parameters in comparison with performance by the WT. The rate of photosynthesis (Pn), which indicates the assimilation efficiency of CO2, was sharply decreased when irrigation was withheld, albeit not to the same extent in the two OE lines as in the WT (Figure 3a). After 8 days of drought treatment, Pn in both OE lines was approximately 1.6 times as high as that in WT plants (Figure 3a). Stomatal conductance followed a similar trend (Figure 3b). Intercellular CO2 concentrations gradually increased during the treatment period, but those increments were smaller in the OE lines (Figure 3c). These gas exchange data suggested that plants overexpressing MdATG18a maintain a better photosynthetic system under drought conditions.
Figure 3.
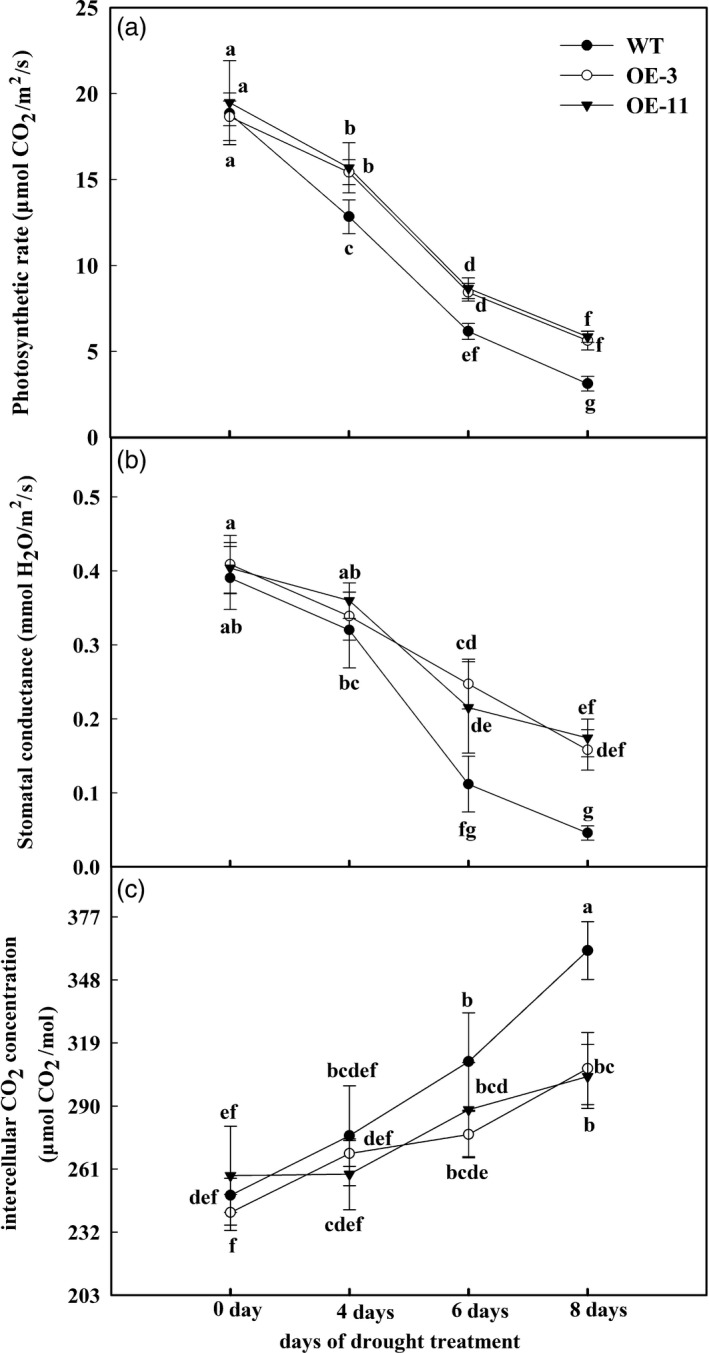
Changes in photosynthesis parameters of MdATG18a‐overexpressing apple relative to untransformed plants during period of drought. (a) Photosynthetic rate, (b) stomatal conductance and (c) intercellular CO 2 concentration. Measurements were made on sunny days between 09:00 and 10:00 h. Data are means of five replicates with SD. Different letters indicate significant differences between treatments, according to one‐way ANOVA Tukey's multiple range tests (P < 0.05).
Apple lines overexpressing MdATG18a accumulate less H2O2 and show enhanced activities of H2O2‐scavenging enzymes under drought stress
As described above, less H2O2 was accumulated in the MdATG18a transgenic tomato plants than in the WT (Figure 1g). To analyse the oxidation status in transgenic apple under drought stress, we measured leaf concentrations of H2O2 and assayed the activities of major antioxidant enzymes. Although 8 days of drought treatment was associated with great accumulations of H2O2 in all plant types, significantly lower amounts were detected in the OE lines (Figure 4a). As the main H2O2‐scavenging enzymes, activities of CAT and POD were obviously increased in response to elevated H2O2 accumulations. For example, when compared with the well‐watered controls, the increases of CAT activity were 1.88 times for OE‐3 and 2.05 times for OE‐11 versus only 1.57 times in the WT (Figure 4b). A similar pattern was observed for POD activities (Figure 4c). Together, these findings indicated that MdATG18a overexpression enhances antioxidant activities in drought‐stressed plants.
Figure 4.
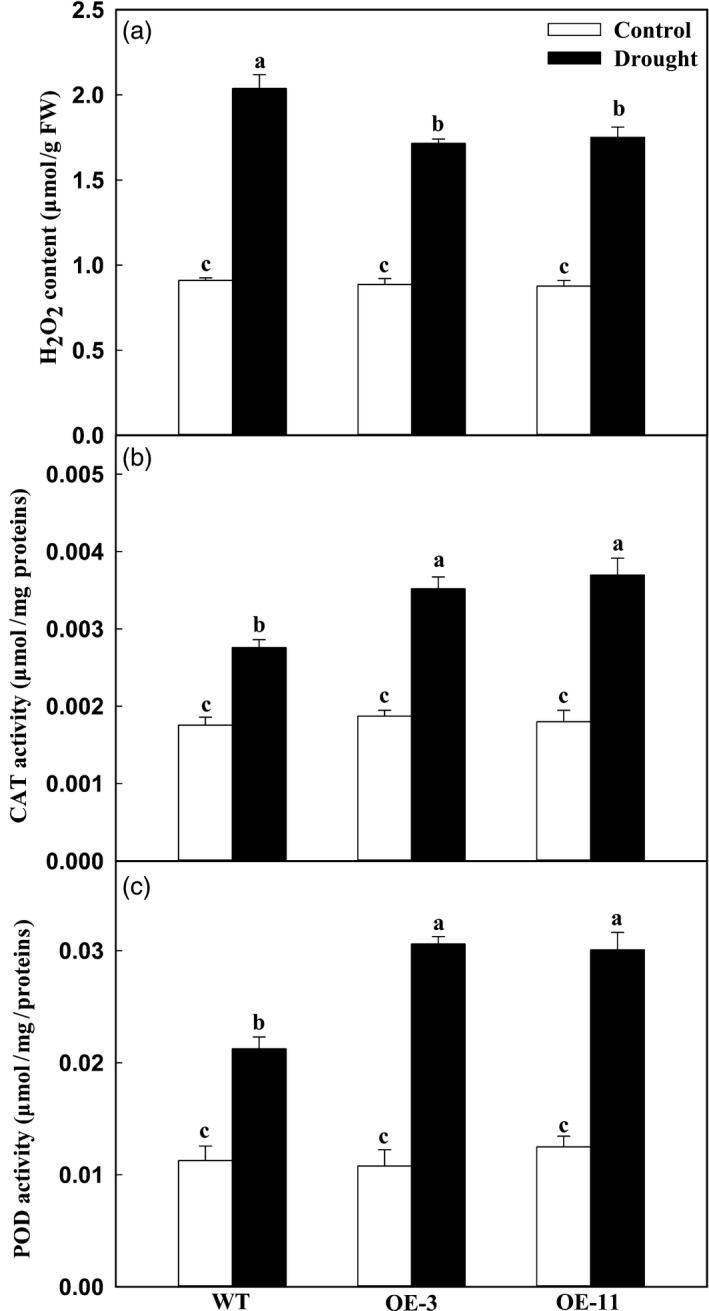
Changes in levels of H2O2 accumulation and activities of ROS‐scavenging enzymes of apple leaves during drought stress. (a) H2O2 concentration. (b) CAT activity. (c) POD activity. These data were measured on Day 8 of drought stress. Data are means of three replicates with SD. Different letters indicate significant differences between treatments, according to one‐way ANOVA Tukey's multiple range tests (P < 0.05).
Apple lines overexpressing MdATG18a show improved AsA‐GSH cycling under drought stress
As an antioxidant system, the AsA–GSH cycle plays an important role in scavenging H2O2 under stress (Wang et al., 2012a). We tested the changes in transcript levels for major genes in that cycle. Under well‐watered conditions, expression of cAPX, MDHAR, DHAR1 and cGR did not differ significantly among genotypes (Figure 5a–d). However, as the drought period became prolonged, transcript levels gradually increased, especially in the transgenic lines. For example, on Day 8 of treatment, expression of cAPX was 3.38 and 3.93 times that of the WT in OE‐3 and OE‐11, respectively (Figure 5a).
Figure 5.
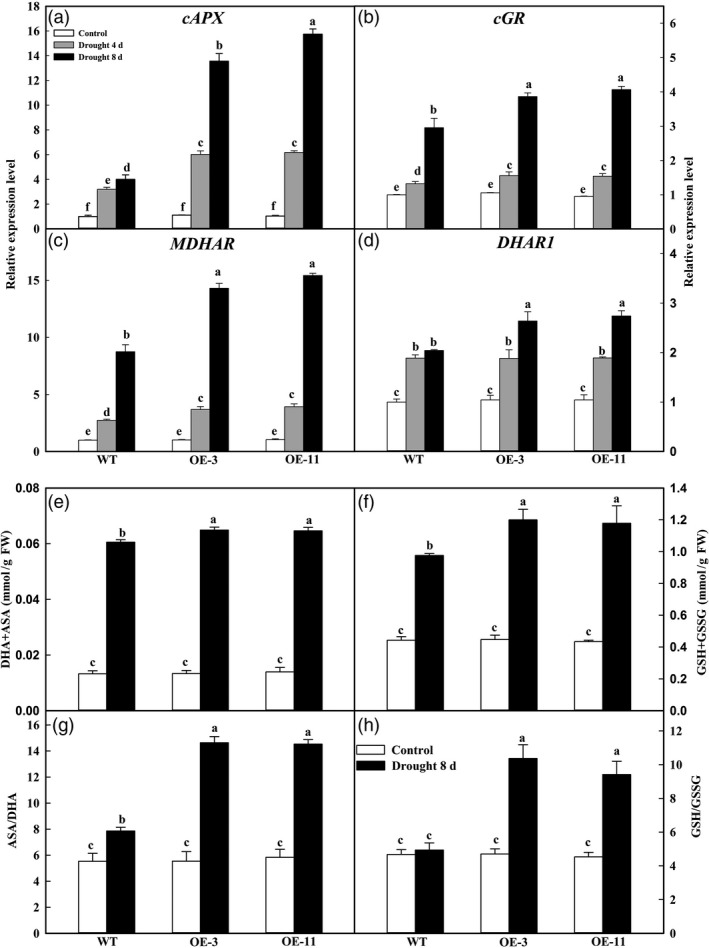
Changes in transcript levels for genes involved in AsA–GSH cycle (a–d) and in concentrations of antioxidants (e–h) of apple leaves during stress period. (a) APX, (b) MDHAR, (c) DHAR and (d) GR, (e) ASA+DHA, (f) ASA/DHA, (g) GSH+GSSG, (h) GSH/GSSG. These data were measured on Day 8 of treatment. Expression levels were calculated relative to expression of Malus EF‐1α mRNA. Data are means of three replicates with SD. Different letters indicate significant differences between treatments, according to one‐way ANOVA Tukey's multiple range tests (P < 0.05).
We further examined the regulation of MdATG18a overexpression in AsA‐GSH cycle by evaluating the states of ascorbate and glutathione. No significant changes in levels of total ascorbate (reduced ascorbate AsA + dehydroascorbate DHA) and total glutathione (reduced glutathione GSH + oxidized glutathione GSSG) were found between the OE lines and WT plants under well‐watered conditions (Figure 5e–h). However, after 8 days of drought treatment, levels of total ascorbates were significantly higher in the transgenics (Figure 5e). This difference was more remarkable in terms of the ratio of AsA to DHA, which was 1.86 and 1.85 times that of the WT in lines OE‐3 and OE‐11, respectively (Figure 5g). Similar results were determined for the total glutathione pool and the GSH/GSSG ratio (Figure 5f,h). Upon induction of drought stress, that ratio did not change in the WT, but increased to 2.11 times for OE‐3 and to 1.75 times for OE‐11 (Figure 5h). Changes in the levels and status of AsA and GSH were in accord with the patterns of transcriptional expression for the main enzymes involved in AsA‐GSH recycling. This clearly demonstrated that overexpressing plants have greater capacity for recycling as well as for maintaining higher amounts of antioxidants.
Apple lines overexpressing MdATG18a accumulate smaller amounts of insoluble and oxidized proteins under drought stress
Autophagy plays a key role in the degradation of oxidized proteins and damaged organelles when plants are stressed (Xiong et al., 2007b). To analyse the relationship between MdATG18a expression and the capacity for such degradation, we measured levels of insoluble and oxidized proteins. In the absence of drought conditions, the amount of insoluble protein as a percentage of the total did not differ between the OE lines and the WT (Figure 6a). However, after 8 days of treatment, insoluble proteins were greatly accumulated in all plants, but those levels were significantly lower in the transgenics. Meanwhile, the proportion of oxidized proteins was also increased by drought in all plants, but accumulations were lower in the OE lines (Figure 6b). These findings strongly suggested that overexpression of MdATG18a can cause oxidized proteins to be degraded and the level of insoluble aggregates to decline when plants are exposed to drought conditions.
Figure 6.
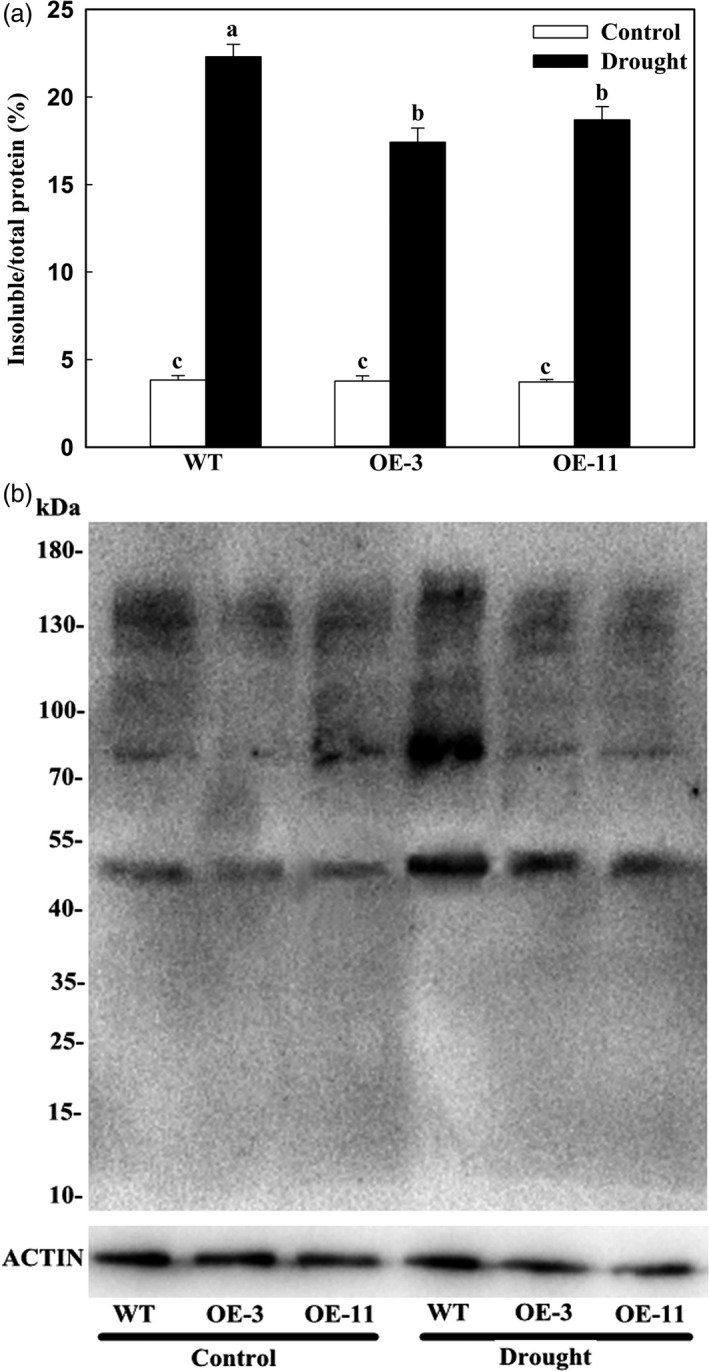
Accumulation of insoluble proteins and oxidation of soluble proteins in apple leaves on Day 8 of drought stress. (a) Accumulation of insoluble proteins in WT and transgenic plants on Day 8 of drought stress. Percentages of insoluble proteins to total proteins were calculated based on amount of total proteins in homogenates at beginning of period and insoluble proteins in final pellets. Data are means of three replicates with SDs. Different letters indicate significant differences between treatments, according to one‐way ANOVA Tukey's multiple range tests (P < 0.05). (b) Oxidation of soluble proteins. Leaf samples were collected after 8 days of treatment, and soluble proteins were isolated and derivatized by 2,4‐dinitrophenol (DNP), followed by immunoblotting with an anti‐DNP antibody. Molecular size markers are indicated at left. Amount of protein extract‐loading was referenced by immunoblot analysis with antiactin antibody (lower panel).
Apple lines overexpressing MdATG18a show up‐regulated expressions of other MdATGs and increased formation of autophagosomes under drought stress
To investigate the occurrence of autophagy in response to drought stress, we examined the expression patterns of several important ATG genes and compared the numbers of autophagosomes that formed in the transgenic lines and WT plants. Expression of MdATG8i, MdATG9 and MdATG10 was significantly higher in OE plants than in the WT, even under well‐watered control conditions (Figure 7). On days 4 and 8 of drought treatment, those three genes, as well as MdATG3a, MdATG5, MdATG7a, MdATG7b, MdATG8c and MdATG8f, were up‐regulated in all genotypes but transcript levels were significantly greater in the OE lines than in the WT.
Figure 7.
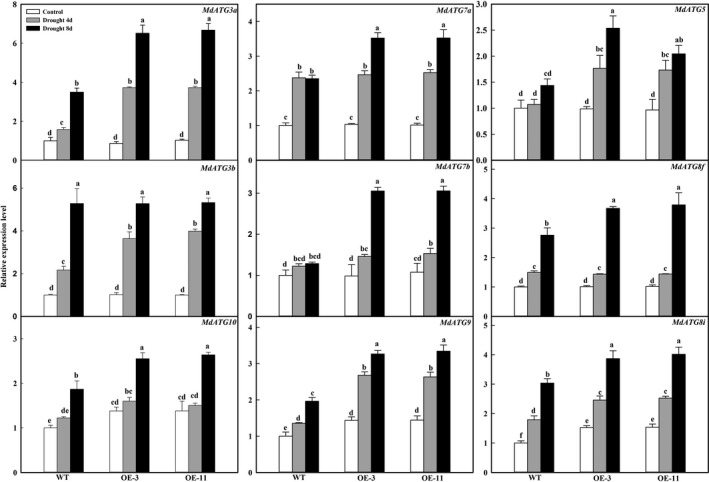
Changes in transcription level of apple autophagy‐related genes during drought period. Total RNA was isolated from leaf samples collected at indicated times, and expression levels were calculated relative to expression of Malus EF‐1α mRNA. Data are means of three replicates with SD. Different letters indicate significant differences between treatments, according to one‐way ANOVA Tukey's multiple range tests (P < 0.05).
We used transmission electron microscopy (TEM) to observe autophagosome formation in response to drought and found very few autophagosome structures in the leaves under well‐watered conditions, regardless of plant type (Figure 8a). However, after 6 days of treatment, up to three times as many autophagosomes and autophagic bodies had accumulated in the OE lines than in WT plants (Figure 8a,b). This demonstrated that the occurrence of autophagy in apple is significantly enhanced by overexpression of MdATG18a when plants are challenged by drought stress.
Figure 8.
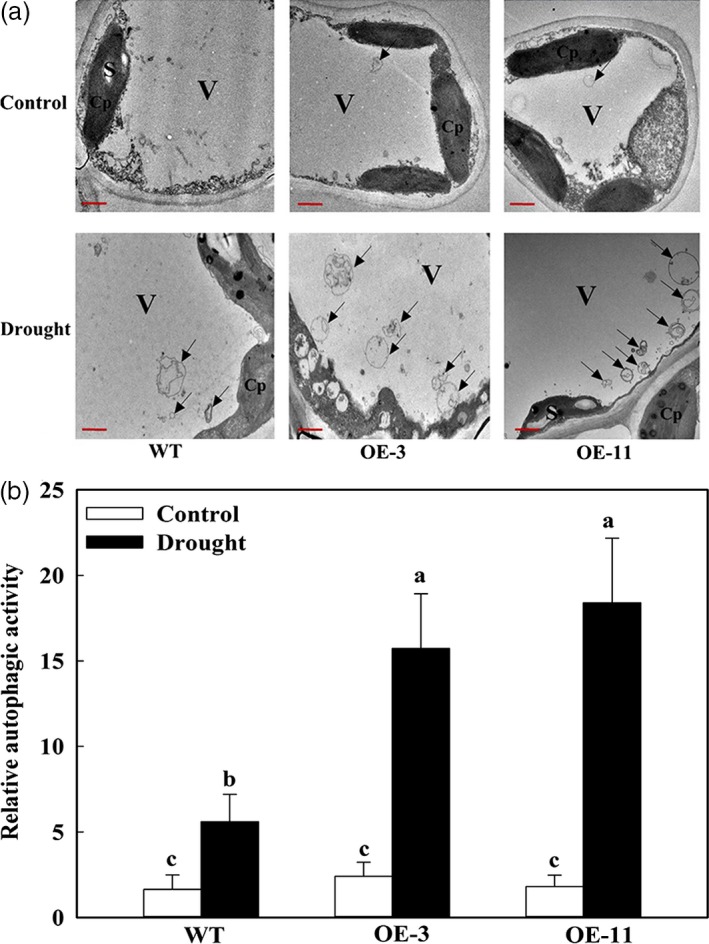
Visualizing the accumulation of autophagosomes in apple leaves under drought stress. (a) Representative TEM images of autophagic structures in mesophyll cells from WT and MdATG18a OE plants. V, vacuole; S, starch; Cp, chloroplast. Autophagic bodies are indicated by black arrows. Bars: 1 μm. (b) Relative autophagic activity normalized to activity of WT or MdATG18a OE plants shown in (a). More than 10 cells were used to quantify structures. Data are means of three replicates with SD. Different letters indicate significant differences between treatments, according to one‐way ANOVA Tukey's multiple range tests (P < 0.05).
Discussion
Autophagy is a conserved cellular degradation process in eukaryotes for survival during environmental stresses and for cellular remodelling during development (Lai et al., 2011; Xiong et al., 2007b). We previously cloned MdATG18a gene from Malus domestica (Wang et al., 2014). Sequence analyses indicated that MdATG18a has typical WD40‐repeat domains which usually serve as scaffolds or platforms for protein–protein interactions and the assembly of protein complexes (Smith et al., 1999). WD40‐repeat proteins are relatively abundant in eukaryotes and are implicated in various important functions including signalling transduction, transcriptional regulation, cell cycle control, apoptosis and autophagy (Nair et al., 2010). MdATG18a showed obvious up‐regulation under leaf senescence and drought stress (Wang et al., 2014). Herein, we used MdATG18a overexpressing plants of tomato and apple to gain more insight into its function and mechanism associated with improved drought tolerance. Our overall data clearly demonstrated that these transgenic plants have improved tolerant to drought stress. Upon drought stress compared with WT, the apple transgenic plants have less damage indicated by parameters of RWC, MDA levels, ROS levels and electrolyte leakage and maintain higher chlorophyll contents and photosynthesis rates, as well as antioxidant ability, which is evidenced by increased activities of H2O2‐scavenging enzymes CAT and POD and greater capacity for AsA‐GSH recycling. Most importantly, there were less oxidized proteins aggregated in MdATG18a OE apple lines, which might be explained by the up‐regulated expressions of other MdATGs and increased formation of autophagosomes under drought stress.
When plants are subjected to drought conditions, RWC values tend to decline and are used as an important indicator of the capacity for water retention (Wang et al., 2012b). We found here that RWC was significantly higher in the transgenic tomato and apple plants than in the WT. Changes in MDA levels and electrolyte leakage in response to drought are two other efficient indicators when evaluating membrane integrity and the extent of tolerance to abiotic stress (Liao et al., 2016; Tang et al., 2013). Here, MDA concentrations and electrolyte leakage were significantly higher in WT tomato and apple plants than in their corresponding transgenics when exposed to drought conditions. This demonstrated that cell damage is alleviated by the overexpression of MdATG18a. The onset of abiotic stress leads to increased production of ROS, causing oxidative damage to cellular components (Kasukabe et al., 2004). Our transgenic tomato plants accumulated less H2O2 and than did the WT after 21 days of treatment. We noted a similar trend in the detection of H2O2 in transgenic apple lines.
Plants utilize both enzymatic and nonenzymatic mechanisms for scavenging ROS (Cheng et al., 2016; Shigeoka et al., 2002; Wang et al., 2015a; Zhou et al., 2013). For example, under drought treatment, overexpression of VqbZIP39 in Arabidopsis increases the activities of CAT, POD and SOD (Tu et al., 2016). Overexpression of MdcyMDH also enhances SOD and CAT activities in transgenic apple callus and in transformed tomato plants (Yao et al., 2011). Likewise, we found that activities of CAT and POD were significantly higher in transgenic apple than in the WT under drought stress. As the most important scavenger of H2O2 in green tissues (Shigeoka et al., 2002), APX is located in different cellular compartments and works in conjunction with the AsA–GSH cycle. Overexpression of MdATG18a up‐regulated the expression of cAPX, as well as MDHAR, DHAR and cGR in that cycling system (Figure 5a–d). The trend in AsA and GSH concentrations and interconversions between their reduced and oxidized forms also fit with the pattern of transcriptional changes we noted in cycling. The levels of total AsA and total GSH, as well as the ratios of AsA/DHA and GSH/GSSG, were significantly elevated in MdATG18a transgenic apple under drought stress. Strong maintenance of a reduced AsA state might be due to the high expression levels of MDHAR and DHAR, which are involved in AsA recycling (Conklin and Barth, 2004). This drought tolerance improvement by MdATG18a overexpression is in accordance with a recent report about transgenic apple plants overexpressing MdcyMDH, which show greater tolerance to salt and cold stresses because the redox state is modified by increases in AsA and GSH levels as well as a reduction in their oxidized ratios (Wang et al., 2016). Drought‐related damage is presumably modulated in MdATG18a transgenic plants because of improvements in their antioxidant systems.
As one of the key processes in primary metabolism, photosynthesis can be affected when water deficits lead to stomatal closure (Chaves et al., 2009). Abiotic stress‐generated ROS can damage the photosynthetic apparatus and inhibit PSII repair because of an imbalance in the redox system in chloroplasts (Gururani et al., 2015). Intracellular ROS accumulation and its inhibition on photosynthesis can be reduced by engineering the production of ROS‐scavenging enzymes such as CAT and APX and by increasing the levels of antioxidants such as AsA and GSH (Gururani et al., 2015). The enhancement of Pn that we found in transgenic apple plants might be a result of an improved antioxidant system under drought stress. Autophagy has a critical role in chloroplast degradation (Wada et al., 2008) and might constitute a dedicated and dynamic quality control mechanism of chloroplasts. However, little is known about how autophagy directly regulates the photosynthetic apparatus or helps in repairing important proteins such as D1 protein.
Autophagy is thought to be involved in degrading oxidized proteins and regulating ROS levels under osmotic or salt stress (Xiong et al., 2007b). It can remove misfolded and damaged proteins or protein aggregates as a mechanism for protein quality control under various stress conditions, such as heat (Yang et al., 2016; Zhou et al., 2014a). Arabidopsis atg5, atg7 and nbr1 mutants have compromised heat tolerance because the plants accumulate insoluble and highly ubiquitinated proteins (Zhou et al., 2014a). Heat stress induces the expression of ATG genes and the accumulation of autophagosomes in tomato plants. Their tolerance to such stress is thought to be mediated by cooperative regulation by both WRKY33 and ATG proteins, probably through the removal of heat‐induced protein aggregates (Zhou et al., 2014a). In tomato, HsfA1a confers drought tolerance through autophagy activation, by which ubiquitinated protein aggregates are degraded in response to stress (Wang et al., 2015b). We also found that insoluble and oxidized proteins were accumulated under drought conditions. However, overexpression of MdATG18a in apple reduced those stress‐related accumulations. The lower amounts of oxidized proteins measured in our OE lines might be explained by activation of autophagy via MdATG18a overexpression. This is supported by the fact that major MdATG genes were significantly up‐regulated and autophagosomes formed in large numbers in the transgenic plants.
Use of TEM is a valid and important method for the quantitative analysis of autophagosomes (Cheng et al., 2016; Klionsky et al., 2012). Our transgenic lines had more autophagosomes than WT under drought stress, a finding that was in line with the decreased accumulation of oxidized insoluble proteins under drought stress. These data again suggest that MdATG18a plays a positive role in removing misfolded and oxidized proteins in drought‐stressed apple, probably through the activation of autophagy.
In conclusion, we have functionally characterized MdATG18a by overexpressing it in tomato and apple. Transgenic apple plants had enhanced drought tolerance, possibly because of a more‐reductive redox state. We propose that active autophagy, as demonstrated by the up‐regulation of autophagy‐related genes and greater autophagosome accumulations in transgenic plants, might contribute to better quality control of proteins and a balanced antioxidant environment under drought stress. Our discoveries provide an interesting link between autophagy and ROS‐scavenging systems, and they show that MdATG18a functions positively in apple drought tolerance. This is a promising perspective for future efforts in crop breeding.
Experimental procedures
Plant materials and treatments
Seeds of Solanum lycopersicum cv. ‘Micro‐Tom’ from transgenic and WT plants were germinated in 250‐cm3 plastic pots in a controlled environment walk‐in chamber. Growing conditions for these tomato plants included 25 °C, 140 μmol photons per m2 per s, 70% relative humidity and a long‐day (14‐h) photoperiod. The seedlings were watered regularly and supplied with half‐strength Hoagland's nutrient solution (pH 6.0) once a week for 20 days to maintain healthy growth. To induce drought stress, some were then exposed to water deprivation by withholding water by 21 days. After drought treatment, the seedlings were rehydrated for 4 days to recover, and those plants with upper leaves turning alive were considered as survived (Zhu et al., 2015). As the well‐watered control group, other plants continued to receive normal irrigation. Samples were harvested on days 0, 14 and 21 of this experimental period, collecting three biological replicates per treatment.
Tissue‐cultured plants of Malus domestica cv. ‘Roya Gala’ were initially grown on an MS agar medium containing 0.3 mg/L 6‐BA and 0.2 mg/L IAA. They were cultured under conditions of 23 °C, 60 μmol/m2/s and a 14‐h photoperiod. After rooting on MS agar media containing 0.5 mg/L IBA and 0.5 mg/L IAA, the transgenic and WT plantlets were transferred to small plastic pots (8.5 × 8.5 × 7.5 cm) containing a mixture of soil/perlite (1 : 1, v : v). After one month of adaptation in a growth chamber, the plants were moved to large plastic pots (30 × 26 × 22 cm) filled with a mixture of forest soil/sand/organic substrate (5 :1 : 1, v : v : v) and grown in the glasshouse. They were watered regularly and supplied with half‐strength Hoagland's nutrient solution (pH 6.0) once a week. After three months of growth under these conditions, healthy and uniformly sized plants were assigned to two treatment groups. Half were subjected to drought stress by withholding water, while the other (well‐watered control) continued to receive daily irrigation so that a saturated soil water content was maintained. On days 0, 4, 6 and 8 of this experiment, between 10:00 and 11:00 h, the ninth to twelfth leaves from the base of a stem (fully mature leaves) were sampled from five trees per treatment. They were pooled together and divided into three repeats. After being rapidly frozen in liquid nitrogen, they were stored at −80 °C.
Construction of plant‐overexpressing vector for MdATG18a
The coding region of MdATG18a was introduced into the pCambia2300 vector by XbaI and KpnI. Primers with restriction sites are listed in Table 1. This vector was driven by the CaMV 35S promoter and carried the kanamycin (Kan) selectable marker in plants. Sequencing‐confirmed plasmid was transformed to Agrobacterium EHA105 by electroporation (Hood et al., 1984).
Table 1.
Primers used in this study
| Name/Accession no. | Sequence (5′‐3′) | Purpose |
|---|---|---|
| oeATG18a | F: GCTCTAGAATGGCCACCCTCTCCGC | Vector construction for plant transformation |
| R:GGGGTACCTTAAAAGGCTTCTTCGGGCTTTA | ||
| qATG18a/ KC800804 | F: ATGATTCCAGGCTTGCCTGCTTTG | Quantitative expression of MdATG18a |
| R: TGCAGCAAAGTTCCGTCGAGAGTA | ||
| EF‐1α/ DQ341381 | F: ATTCAAGTATGCCTGGGTGC | Real‐time PCR using Malus EF‐1α as reference gene |
| R: CAGTCAGCCTGTGATGTTCC | ||
| dATG18a | F: GAGAACACGGGGGACTCTAGA | DNA confirmation of MdATG18a for transgenic identification |
| R: CGATCGGGGAAATTCGAGCTC | ||
| qcAPX/ EF528482 | F: AACTACAAGGGATGAAGCC | Quantitative expression of MdcAPX |
| R: CAACGAGGATGATAACCAG | ||
| qMDHAR/ FJ752239 | F: CCATACTTCTATTCCCGCTCCT | Quantitative expression of MdMDHAR |
| R: CGACCACCTTCCCGTCTTT | ||
| qDHAR1/ DQ322706 | F: AGTGGACGGTTCCAGCAGA | Quantitative expression of MdDHAR1 |
| R: AGTGGACGGTTCCAGCAGA | ||
| qGR/ JF268781 | F: GTTCAGCGACAAGGCGTAT | Quantitative expression of MdGR |
| R: TCAACCGATTTCCATTTCC | ||
| qATG3a/ KF438032 | F: AAGGGGGCGGAGATGGTTC | Quantitative expression of MdATG3a |
| R: GCACTTAGAGACGAGGTTATCGC | ||
| qATG3b/ KR024682 | F: AGGGAGATGGTTTTGAAACAGA | Quantitative expression of MdATG3b |
| R: ACTTAGAGACGAGGTTATCGC | ||
| qATG5/ KY305671 | F: GCAGGTCGTGTTCCAGTTC | Quantitative expression of MdATG5 |
| R: CCTCCTCCTCCTTGTATCTCAA | ||
| qATG7a/ KF438034 | F: GCGGATATGAGCAACCTTGGC | Quantitative expression of MdATG7a |
| R: ATCAATAGGCGCAACGACATCA | ||
| qATG7b/ KF438035 | F: ATCGGTAACAGGAGTAAGTCGG | Quantitative expression of MdATG7b |
| R: TTTATCAAGCGCATGAAAGCCT | ||
| qATG8f/ KF438036 | F: TCGTAGACAATGTCCTCCCAGC | Quantitative expression of MdATG8f |
| R: CCAAATGTGTTCTCGCCACTGT | ||
| qATG8i/ KF438037 | F: GCAGCAGGCTTCACTTGACTCC | Quantitative expression of MdATG8i |
| R: GGAATCCATGCGACTGGCTGTT | ||
| qATG9/ KF438038 | F: ACTTCATGCGTCAGCCTTCAGA | Quantitative expression of MdATG9 |
| R: CGTTCCTCCAATCCAACCGTTG | ||
| qATG10/ KF438033 | F: TGGAACCAGCGAGTGGATGAAG | Quantitative expression of MdATG10 |
| R: ACAACTGAGAGCCAAGACACCA | ||
| Actin/Sl11g005330 | F: TGTCCCTATTTACGAGGGTTATGC | Real‐time PCR of actin as a reference gene in tomato |
| R: CAGTTAAATCACGACCAGCAAGAT |
Genetic transformation of tomato and apple
‘Micro‐Tom’ tomato was transformed via Agrobacterium‐mediated method, as previously described (Guo et al., 2012). The resultant plants were PCR‐confirmed to select positive transgenic lines. Nine transgenic lines were obtained at the beginning, and they were kept screening with 100 mg/L Kan. OE‐1 and OE‐9 were two lines that did not segregate anymore until F2 generation. Meanwhile, Southern blot checked the copy numbers of these lines and confirmed the single‐copy insertions in OE‐1 and OE‐9 lines. Therefore, these two homozygous lines were selected for further tests.
Transgenic ‘Royal Gala’ apple plants were generated from leaf fragments through Agrobacterium‐mediated transformation, as previously described (Dai et al., 2013). Regenerated Kan‐resistant buds were subcultured every three weeks, based on selection with 25 mg/L Kan. Lines that stopped growing or died were eliminated and only those showing normal growth were maintained for further verification. We performed PCR analysis with isolated DNA to check for the presence of the transgene in putative transformed lines. Plasmid and nontransformed plant DNA were used as the positive and negative controls, respectively. Total RNA was then isolated from positive lines as well as from the nontransformed WT according to a CTAB method (Chang et al., 1993). Overexpression of MdATG18a was confirmed by quantitative real‐time PCR (qRT–PCR).
Southern blot
The PCR‐positive transgenic tomato and apple lines were analysed for insertions by Southern blotting. Primers (forward: AGATCCTCGCCGTCGGGCATG and reverse: AGATCCTCGCCGTCGGGCATG) were used to amplify the NPTII fragment to prepare the labelled probe using the PCR DIG Probe Synthesis Kit (Roche, Mannheim, Switzerland). Genomic DNA was extracted from fresh young leaves of every transgenic tomato and apple line and nontransgenic controls. Approximately 40 μg of genomic DNA from each genotype was used and completely digested with the restriction enzyme EcoRI, and the plasmid was digested with the restriction enzyme XbaI. Southern blotting was performed according to the DIG DNA Labeling and Detection Kit (Roche, Switzerland) manual instructions.
RNA extraction, DNA isolation and qRT–PCR
Total RNA was extracted according to a CTAB method (Chang et al., 1993). The DNA was removed by treating with RNase‐free DNase I (Thermo Scientific, Waltham, MA). Apple genomic DNA was extracted by a modified CTAB method (Modgil et al., 2005), and first‐strand cDNA was synthesized using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) with the same amount of mRNA (1 μg). qRT–PCR was performed with the iQ5 Multicolor Real‐Time PCR Detection System (Bio‐Rad Laboratories, Hercules, CA) and SYBR Green Master Mix (Takara, Dalian, China). Transcripts of the Malus elongation factor 1 alpha gene (EF‐1α; DQ341381) were used to standardize the cDNA samples for different genes. Specific primer sequences for expression analysis are shown in Table 1. All experiments were repeated three times biologically, based on three separate RNA extracts from three repeats.
Measurements of gas exchange parameters
Gas exchange parameters were monitored by a LI‐COR 6400 portable photosynthesis system (LI‐COR, Huntington Beach, CA). Measurements were performed on the ninth to twelfth leaves from the base of selected plant stems on sunny days between 09:00 and 10:00 h. All photosynthetic measurements were recorded at a constant airflow rate of 500 μmol/s. The concentration of CO2 was 400 ± 5 cm3/m3, and the temperature was 28 ± 2 °C. The rate of photosynthesis (Pn), intercellular CO2 concentration and stomatal conductance were obtained from five plants per treatment. Measurements were made at a photosynthetic photon flux density of 1000 μmol/m2/s, as provided by a Q‐Beam (blue and red diode) light source.
Evaluation of stress tolerance
Relative water contents were determined as previously described (Gaxiola et al., 2001). Briefly, fresh weights (FWs) of the terminal leaflets were measured before the leaves were immersed in distilled water under darkness for 24 h to obtain their fully turgid weights (TWs). They were then oven‐dried at 70 °C for 72 h before their dry weights (DWs) were recorded. The relative water content was calculated as follows:
Chlorophyll was extracted with 80% acetone, and concentrations were determined spectrophotometrically according to the method of Lichtenthaler and Wellburn (1983). Levels of MDA were obtained as previously described (Heath and Packer, 1968). Electrolyte leakage in the leaves was calculated according to an earlier method (Thalhammer et al., 2014). Finally, H2O2 was extracted with 5% (w/v) trichloroacetic acid and measured as described previously (Patterson et al., 1984).
Detection of H2O2 and superoxide ion ()
Accumulations of and H2O2 were examined by histochemical staining methods that used nitro blue tetrazolium (NBT) and diaminobenzidine (DAB), respectively. For detection, leaves were incubated under darkness for 4 h at 25 °C in fresh NBT solution (1 mg/mL) prepared in 10 mm Hepes (pH 7.5) with 0.1% Triton X‐100. For H2O2 detection, leaves were placed in a fresh DAB (1 mg/mL) solution prepared in 10 mm sodium phosphate buffer (pH 7.5) with 0.1% Triton X‐100. The samples were then incubated in a growth chamber overnight until brown spots were visible. The chlorophyll was removed by immersing the leaves in 90% ethanol and heating in a boiling water‐bath for 15 min. Samples were fixed in 90% ethanol with 20% glycerol at 4 °C prior to photographing (Rangani et al., 2016; Wang et al., 2015a).
Extraction and assay of antioxidant enzymes
Apple leaves (0.1 g) were ground with a 1.2 mL ice‐cold buffer containing 50 mm potassium phosphate buffer (pH 7.8), 1 mm EDTA‐Na2, 0.3% Triton X‐100 and 1% (w/v) polyvinylpolypyrrolidone. The homogenate was centrifuged at 13 000 g for 20 min at 4 °C, and the supernatant was used for the assays. Activities of CAT and POD were determined according to established protocols (Wang et al., 2012a; Zhang et al., 2015).
Extraction and analysis of antioxidant metabolites
Both AsA and DHA were extracted with 6% (v/v) HClO4, while GSH and GSSG were extracted with 5% (v/v) sulfosalicylic acid. Their concentrations were measured as previously described (Wang et al., 2012a).
Detection of insoluble proteins and Western blotting
Soluble, insoluble and total proteins were measured according to earlier methods (Zhou et al., 2013). Their concentrations were determined with protein assay kits (Bio‐Rad), using bovine serum albumin as a standard. Oxidized proteins from the soluble protein fraction were detected with an OxyBlot protein oxidation detection kit (Chemicon International, Temecula, CA), based on the manufacturer's instructions. Actin was monitored with a monoclonal antibody (CWBIO, Beijing, China). After incubation with a horseradish peroxidase‐linked secondary antibody (CWBIO), the antigen–antibody complexes were detected using Clarity™ Western ECL Substrate (Bio‐Rad) according to the manufacturer's instructions.
Transmission electron microscopy analysis
Our TEM analysis was performed as previously described, but with slight modifications (Wang et al., 2015b). On Day 6 of the drought stress period, mature leaves were excised from the apple plants and immediately cut into small pieces and then fixed with 2.5% glutaraldehyde in 0.2 m PBS buffer (pH 7.4) before being placed under darkness for 12 h at 4 °C. After washes with PBS buffer, the samples were fixed for 2.5 h in 1% (v/v) osmium tetroxide at room temperature. They were then dehydrated in a graded ethanol series (30%–100%; v/v) and embedded in Epon 812. Ultrathin sections (70 nm) were prepared on an ultramicrotome (Leica ULTRACUT, Wetzlar, Germany) and collected on Formvar‐coated grids. The sections were examined using a JEOL‐1230 transmission electron microscope (Hitachi, Tokyo, Japan) at an accelerating voltage of 80 kV to observe autophagosomes and autophagic bodies.
Statistical analysis
Three independent replicates were used for each determination. Experimental data were presented as means ± standard deviation (SD). Statistical data analysis was performed via one‐way ANOVA, followed by Tukey's multiple range tests, using the SPSS 18 statistical package SPSS, Chicago, Illinois, USA. Differences among results were considered statistically significant at P < 0.05.
Conflict of interest
The authors declare no conflicts of interest.
Supporting information
Figure S1 Southern blot of MdATG18a transgenic lines of tomato and apple.
Acknowledgements
This work was supported by the State Key Program of the National Natural Science Foundation of China (31330068), the Young Scientists Fund of the National Natural Science Foundation of China (31601735) and by the earmarked fund for the China Agriculture Research System (CARS‐28). The authors are grateful to Dr. Zhihong Zhang for providing tissue‐cultured apple plants, to Priscilla Licht for help in revising our English composition and to Mr. Zhengwei Ma for management of the apple trees.
References
- Apel, K. and Hirt, H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Barth, H. , Meiling‐Wesse, K. , Epple, U.D. and Thumm, M. (2001) Autophagy and the cytoplasm to vacuole targeting pathway both require Aut10p. FEBS Lett. 508, 23–28. [DOI] [PubMed] [Google Scholar]
- Chang, S. , Puryear, J. and Cairney, J. (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. [Google Scholar]
- Chaves, M.M. , Flexas, J. and Pinheiro, C. (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann. Bot. 103, 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Liao, B. , Qi, H. , Xie, L.J. , Huang, L. , Tan, W.J. , Zhai, N. et al (2015) Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana . Autophagy, 11, 2233–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, F. , Yin, L.L. , Zhou, J. , Xia, X.J. , Shi, K. , Yu, J.Q. , Zhou, Y.H. et al (2016) Interactions between 2‐Cys peroxiredoxins and ascorbate in autophagosome formation during the heat stress response in Solanum lycopersicum . J. Exp. Bot. 67, 1919–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin, P. and Barth, C. (2004) Ascorbic acid, a familiar small molecule intertwined in the response of plants to ozone, pathogens, and the onset of senescence. Plant, Cell Environ. 27, 959–970. [Google Scholar]
- Dai, H. , Li, W. , Han, G. , Yang, Y. , Ma, Y. , Li, H. and Zhang, Z. (2013) Development of a seedling clone with high regeneration capacity and susceptibility to Agrobacterium in apple. Sci. Hortic. 164, 202–208. [Google Scholar]
- Foyer, C.H. and Noctor, G. (2009) Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid. Redox Signal. 11, 861–905. [DOI] [PubMed] [Google Scholar]
- Gaxiola, R.A. , Li, J. , Undurraga, S. , Dang, L.M. , Allen, G.J. , Alper, S.L. and Fink, G.R. (2001) Drought‐and salt‐tolerant plants result from overexpression of the AVP1 H+‐pump. Proc. Natl Acad. Sci. USA, 98, 11444–11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, J. , Stromhaug, P.E. , George, M.D. , Habibzadegah‐Tari, P. , Bevan, A. , Dunn, W.A. and Klionsky, D.J. (2001) Cvt18/Gsa12 is required for cytoplasm‐to‐vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris . Mol. Biol. Cell, 12, 3821–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M. , Zhang, Y. , Meng, Z. and Jiang, J. (2012) Optimization of factors affecting Agrobacterium‐mediated transformation of Micro‐Tom tomatoes. Genet. Mol. Res. 11, 661–671. [DOI] [PubMed] [Google Scholar]
- Gururani, M.A. , Venkatesh, J. and Tran, L.S.P. (2015) Regulation of photosynthesis during abiotic stress‐induced photoinhibition. Mol. Plant. 8, 1304–1320. [DOI] [PubMed] [Google Scholar]
- Han, S. , Yu, B. , Wang, Y. and Liu, Y. (2011) Role of plant autophagy in stress response. Protein Cell, 2, 784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, S. , Wang, Y. , Zheng, X. , Jia, Q. , Zhao, J. , Bai, F. , Hong, Y. et al (2015) Cytoplastic glyceraldehyde‐3‐phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana . Plant Cell, 27, 1316–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havé, M. , Marmagne, A. , Chardon, F. and Masclaux‐Daubresse, C. (2016) Nitrogen remobilisation during leaf senescence: lessons from Arabidopsis to crops. J. Exp. Bot. 68, 2513–2529. [DOI] [PubMed] [Google Scholar]
- Heath, R.L. and Packer, L. (1968) Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198. [DOI] [PubMed] [Google Scholar]
- Hood, E.E. , Jen, G. , Kayes, L. , Kramer, J. , Fraley, R.T. and Chilton, M.D. (1984) Restriction endonuclease map of pTi Bo542, a potential Ti plasmid vector for genetic engineering of plants. Nat. Biotechnol. 2, 702–709. [Google Scholar]
- Kasukabe, Y. , He, L. , Nada, K. , Misawa, S. , Ihara, I. and Tachibana, S. (2004) Overexpression of spermidine synthase enhances tolerance to multiple environmental stresses and up‐regulates the expression of various stress‐regulated genes in transgenic Arabidopsis thaliana . Plant Cell Physiol. 45, 712–722. [DOI] [PubMed] [Google Scholar]
- Klionsky, D.J. , Abdalla, F.C. , Abeliovich, H. , Abraham, R.T. , Acevedo‐Arozena, A. , Adeli, K. , Agholme, L. et al (2012) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy, 8, 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick, R. , Tolstrup, J. , Appelles, A. , Henke, S. and Thumm, M. (2006) The relevance of the phosphatidylinositolphosphat‐binding motif FRRGT of Atg18 and Atg21 for the Cvt pathway and autophagy. FEBS Lett. 580, 4632–4638. [DOI] [PubMed] [Google Scholar]
- Lai, Z. , Wang, F. , Zheng, Z. , Fan, B. and Chen, Z. (2011) A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 66, 953–968. [DOI] [PubMed] [Google Scholar]
- Levine, A. , Tenhaken, R. , Dixon, R. and Lamb, C. (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell, 79, 583–593. [DOI] [PubMed] [Google Scholar]
- Liao, X. , Guo, X. , Wang, Q. , Wang, Y. , Zhao, D. , Yao, L. , Wang, S. et al (2016) Overexpression of MsDREB6.2 results in cytokinin‐deficient developmental phenotypes and enhances drought tolerance in transgenic apple plants. Plant J. 8, 510–526. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. and Wellburn, A.R. (1983) Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 11, 591–592. [Google Scholar]
- Liu, Y. and Bassham, D.C. (2012) Autophagy: pathways for self‐eating in plant cells. Annu. Rev. Plant Biol. 63, 215–237. [DOI] [PubMed] [Google Scholar]
- Liu, Y.M. , Xiong, Y. and Bassham, D.C. (2009) Autophagy is required for tolerance of drought and salt stress in plants. Autophagy, 5, 954–963. [DOI] [PubMed] [Google Scholar]
- Mizushima, N. and Komatsu, M. (2011) Autophagy: renovation of cells and tissues. Cell, 147, 728–741. [DOI] [PubMed] [Google Scholar]
- Modgil, M. , Mahajan, K. , Chakrabarti, S. , Sharma, D. and Sobti, R. (2005) Molecular analysis of genetic stability in micropropagated apple rootstock MM106. Sci. Hortic. 104, 151–160. [Google Scholar]
- Nair, U. , Cao, Y. , Xie, Z. and Klionsky, D.J. (2010) Roles of the lipid‐binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J. Biol. Chem. 285, 11476–11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, B.D. , MacRae, E.A. and Ferguson, I.B. (1984) Estimation of hydrogen peroxide in plant extracts using titanium (IV). Anal. Biochem. 139, 487–492. [DOI] [PubMed] [Google Scholar]
- Pompelli, M.F. , Barata‐Luís, R. , Vitorino, H.S. , Gonçalves, E.R. , Rolim, E.V. , Santos, M.G. , Almeida‐Cortez, J.S. et al (2010) Photosynthesis, photoprotection and antioxidant activity of purging nut under drought deficit and recovery. Biomass Bioenerg. 34, 1207–1215. [Google Scholar]
- Rangani, J. , Parida, A.K. , Panda, A. and Kumari, A. (2016) Coordinated changes in antioxidative enzymes protect the photosynthetic machinery from salinity induced oxidative damage and confer salt tolerance in an extreme halophyte Salvadora persica L. Front Plant Sci. 7, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabovol, V. and Minibayeva, F. (2016) Molecular mechanisms of autophagy in plants: Role of ATG8 proteins in formation and functioning of autophagosomes. Biochemistry (Moscow), 81, 348–363. [DOI] [PubMed] [Google Scholar]
- Shigeoka, S. , Ishikawa, T. , Tamoi, M. , Miyagawa, Y. , Takeda, T. , Yabuta, Y. and Yoshimura, K. (2002) Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 53, 1305–1319. [PubMed] [Google Scholar]
- Shin, J.H. , Yoshimoto, K. , Ohsumi, Y. , Jeon, J.S. and An, G. (2009) OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol. Cells, 27, 67–74. [DOI] [PubMed] [Google Scholar]
- Smith, T.F. , Gaitatzes, C. , Saxena, K. and Neer, E.J. (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24, 181–185. [DOI] [PubMed] [Google Scholar]
- Tang, L. , Cai, H. , Ji, W. , Luo, X. , Wang, Z. , Wu, J. , Wang, X. et al (2013) Overexpression of GsZFP1 enhances salt and drought tolerance in transgenic alfalfa (Medicago sativa L.). Plant Physiol. Biochem. 71, 22–30. [DOI] [PubMed] [Google Scholar]
- Thalhammer, A. , Hincha, D.K. and Zuther, E. (2014) Measuring freezing tolerance: electrolyte leakage and chlorophyll fluorescence assays In Plant Cold Acclimation: Methods and Protocols (Hincha D.K. and Zuther E., eds), Methods in Molecular Biology, 1166, pp. 15–24. New York: Springer Science+ Business Media. [DOI] [PubMed] [Google Scholar]
- Tsugane, K. , Kobayashi, K. , Niwa, Y. , Ohba, Y. , Wada, K. and Kobayashi, H. (1999) A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell, 11, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, M. , Wang, X. , Huang, L. , Guo, R. , Zhang, H. , Cai, J. and Wang, X. (2016) Expression of a grape bZIP transcription factor, VqbZIP39, in transgenic Arabidopsis thaliana confers tolerance of multiple abiotic stresses. Plant Cell, Tissue Organ Cult. 125, 537–551. [Google Scholar]
- Wada, S. , Ishida, H. , Izumi, M. , Yoshimoto, K. , Ohsumi, Y. , Mae, T. and Makino, A. (2008) Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149, 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Yin, L. , Liang, D. , Li, C. , Ma, F. and Yue, Z. (2012a) Delayed senescence of apple leaves by exogenous melatonin treatment: toward regulating the ascorbate–glutathione cycle. J. Pineal Res. 53, 11–20. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Liang, D. , Li, C. , Hao, Y. , Ma, F. and Shu, H. (2012b) Influence of drought stress on the cellular ultrastructure and antioxidant system in leaves of drought‐tolerant and drought‐sensitive apple rootstocks. Plant Physiol. Biochem. 51, 81–89. [DOI] [PubMed] [Google Scholar]
- Wang, P. , Sun, X. , Yue, Z. , Liang, D. , Wang, N. and Ma, F. (2014) Isolation and characterization of MdATG18a, a WD40‐repeat AuTophaGy‐related gene responsive to leaf senescence and abiotic stress in Malus . Sci. Hortic. 165, 51–61. [Google Scholar]
- Wang, P. , Sun, X. , Wang, N. , Tan, D.X. and Ma, F. (2015a) Melatonin enhances the occurrence of autophagy induced by oxidative stress in Arabidopsis seedlings. J. Pineal Res. 58, 479–489. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Cai, S. , Yin, L. , Shi, K. , Xia, X. , Zhou, Y. , Yu, J. et al (2015b) Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy, 11, 2033–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Sun, H. , Dong, Q. , Sun, T. , Jin, Z. , Hao, Y. and Yao, Y. (2016) The enhancement of tolerance to salt and cold stresses by modifying the redox state and salicylic acid content via the cytosolic malate dehydrogenase gene in transgenic apple plants. Plant Biotechnol. J. 14, 1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, Y. , Contento, A.L. and Bassham, D.C. (2005) AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana . Plant J. 42, 535–546. [DOI] [PubMed] [Google Scholar]
- Xiong, Y. , Contento, A.L. and Bassham, D.C. (2007a) Disruption of autophagy results in constitutive oxidative stress in Arabidopsis . Autophagy, 3, 257–258. [DOI] [PubMed] [Google Scholar]
- Xiong, Y. , Contento, A.L. , Nguyen, P.Q. and Bassham, D.C. (2007b) Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis . Plant Physiol. 143, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Srivastava, R. , Howell, S.H. and Bassham, D.C. (2016) Activation of autophagy by unfolded proteins during endoplasmic reticulum stress. Plant J. 85, 83–95. [DOI] [PubMed] [Google Scholar]
- Yao, Y.X. , Li, M. , Zhai, H. , You, C.X. and Hao, Y.J. (2011) Isolation and characterization of an apple cytosolic malate dehydrogenase gene reveal its function in malate synthesis. J. Plant Physiol. 168, 474–480. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Niu, J. , Duan, Y. , Zhang, M. , Liu, J. , Li, P. and Ma, F. (2015) Photoprotection mechanism in the ‘Fuji’ apple peel at different levels of photooxidative sunburn. Physiol. Plant. 154, 54–65. [DOI] [PubMed] [Google Scholar]
- Zhao, M. and Running, S.W. (2010) Drought‐induced reduction in global terrestrial net primary production from 2000 through 2009. Science, 329, 940–943. [DOI] [PubMed] [Google Scholar]
- Zhou, J. , Wang, J. , Cheng, Y. , Chi, Y.J. , Fan, B. , Yu, J.Q. and Chen, Z. (2013) NBR1‐mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9, e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Wang, J. , Yu, J.Q. and Chen, Z. (2014a) Role and regulation of autophagy in heat stress responses of tomato plants. Front Plant Sci. 5, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Zhang, Y. , Qi, J. , Chi, Y. , Fan, B. , Yu, J.Q. and Chen, Z. (2014b) E3 ubiquitin ligase CHIP and NBR1‐mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 10, e1004116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, M. , Chen, G. , Dong, T. , Wang, L. , Zhang, J. , Zhao, Z. and Hu, Z. (2015) SlDEAD31, a putative DEAD‐Box RNA helicase gene, regulates salt and drought tolerance and stress‐related genes in tomato. PLoS ONE, 10, e0133849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Southern blot of MdATG18a transgenic lines of tomato and apple.


