Figure 5.
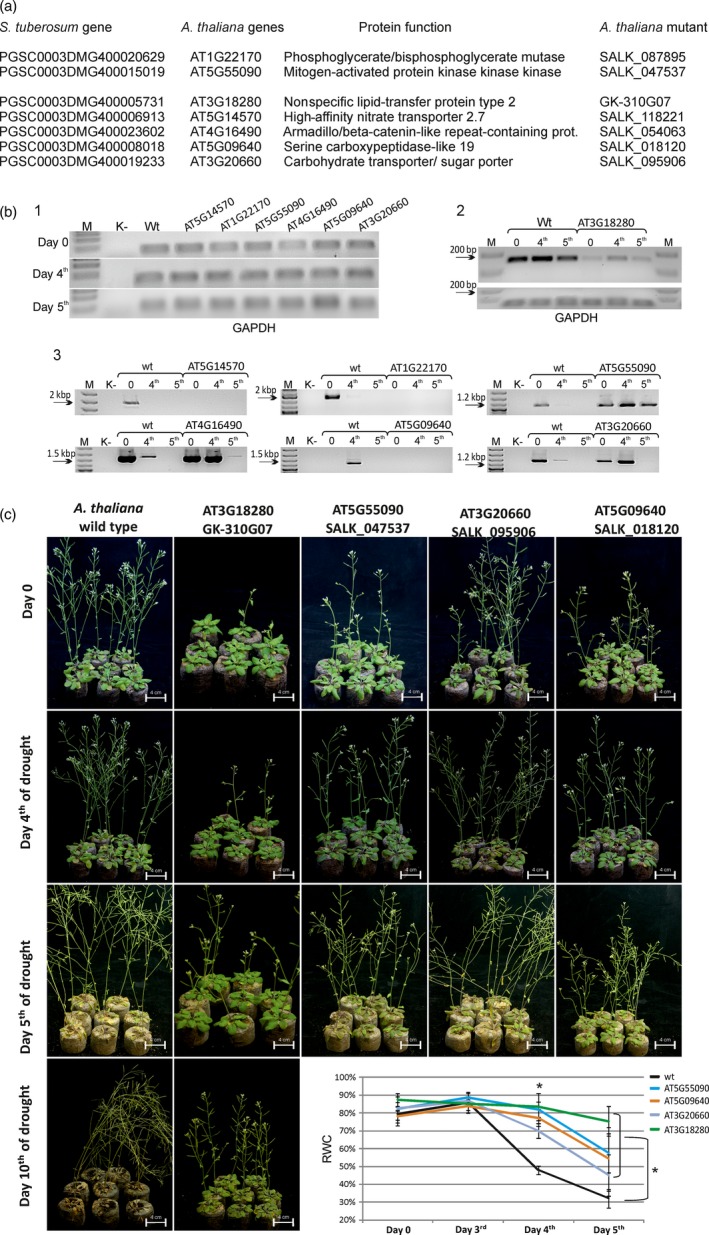
Analysis of Arabidopsis mutant plants with altered expression of genes homologous to the selected drought‐related potato genes. (a) Accession numbers of the seven top‐ranking drought‐related potato genes (left column) and the accession numbers and protein functions of their Arabidopsis homologues (middle columns) were obtained from the Spud DB and TAIR database (Hirsch et al., 2014; Huala et al., 2001). The right column presents the accession numbers of the selected Arabidopsis SALK or GABI‐Kat mutants of each gene (Alonso et al., 2003; Kleinboelting et al., 2012). (b) RT‐PCR analysis of the expression of selected Arabidopsis genes under control (day 0) and drought conditions (days 4 and 5). (1) Gel electrophoresis of the RT‐PCR products of the GAPDH gene in wild‐type and selected Arabidopsis SALK line plants on days 0, 4 and 5 of the drought experiment; (2 and 3) gel electrophoresis of the RT‐PCR products of selected Arabidopsis genes in wild‐type and mutant plants grown under control and drought conditions, respectively. SALK accession numbers are presented above each electrophoretic line. M—DNA marker; K—RT‐PCR nontemplate control; wt—wild‐type plant. (c) Arabidopsis plants subjected to water deficit stress. Plants are shown on days 0, 4, 5 and 10 of the drought experiment. Only wild‐type and four mutant plants are shown (the other three are presented in Fig. S8). Lower bottom right panel of (c)—RWC analysis in the leaves of wild‐type and selected mutant Arabidopsis plants. Mutant plants show higher water content than wild‐type plants. RWC was measured 0, 4 and 5 days after the introduction of drought stress. Values are shown as mean ± SD (n = 3) of three independent experiments. *P < 0.001, Mann–Whitney U‐test.
